Page 303 - Applied Probability
P. 303
13. Sequence Analysis
291
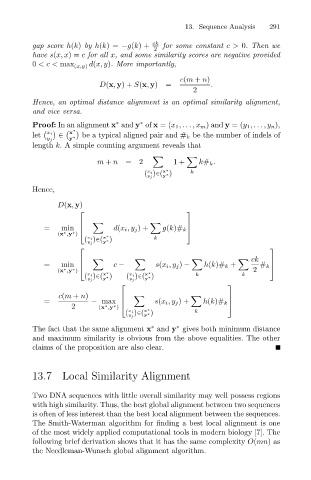
ck
gap score h(k) by h(k)= −g(k)+
2
have s(x, x)= c for all x, and some similarity scores are negative provided
0 <c< max (x,y) d(x, y). More importantly,
c(m + n)
.
=
D(x, y)+ S(x, y) for some constant c> 0. Then we
2
Hence, an optimal distance alignment is an optimal similarity alignment,
and vice versa.
Proof: In an alignment x and y of x =(x 1 ,...,x m ) and y =(y 1 ,...,y n),
∗
∗
x
∗
let x i ∈ y ∗ be a typical aligned pair and # k be the number of indels of
y j
length k. A simple counting argument reveals that
m + n =2 1+ k# k .
x ∗
x i ∈( y ∗) k
)
( y j
Hence,
D(x, y)
= d(x i ,y j )+
min
(x ∗ ,y ∗ ) g(k)# k
x ∗
)
x i ∈( y ∗) k
( y j
ck
= min c − s(x i ,y j ) − h(k)# k + # k
∗
∗
(x ,y ) 2
)
x ∗
x ∗
x i ∈( y ∗) x i ∈( y ∗) k k
)
( y j ( y j
c(m + n)
= s(x i ,y j )+
2 − max h(k)# k
(x ,y )
∗
∗
x ∗
)
x i ∈( y ∗) k
( y j
∗
The fact that the same alignment x and y gives both minimum distance
∗
and maximum similarity is obvious from the above equalities. The other
claims of the proposition are also clear.
13.7 Local Similarity Alignment
Two DNA sequences with little overall similarity may well possess regions
with high similarity. Thus, the best global alignment between two sequences
is often of less interest than the best local alignment between the sequences.
The Smith-Waterman algorithm for finding a best local alignment is one
of the most widely applied computational tools in modern biology [7]. The
following brief derivation shows that it has the same complexity O(mn)as
the Needleman-Wunsch global alignment algorithm.