Page 50 - Cascade biocatalysis
P. 50
26 2 New Trends in the In Situ Enzymatic Recycling of NAD(P)(H) Cofactors
into the organic solvent, whereas the enzymes, the cofactor and, particularly,
the co-substrate glucose, and the co-product gluconate are dissolved in the
aqueous buffer [21–23]. Moreover, this solution provides the additional advantage
of minimizing possible substrate/product inhibition effects and it gives the
opportunity to reuse the enzymes simply by separating the two phases and
supplying a fresh substrate solution.
2.2.1.2 Phosphite Dehydrogenase
A very promising NAD(P)H regenerating system is that which is based on the
oxidation of phosphite to phosphate, catalyzed by phosphite dehydrogenase (PTDH,
EC 1.20.1.1, Scheme 2.1), because of its highly favorable equilibrium constant
+
around 10 11 for NAD reduction [24–26]. The enzyme was first characterized
from a Pseudomonas stutzeri strain, actually showing a k cat for NADH oxidation
only slightly higher than that of C. boidinii FDH. Moreover, the wild-type PTDH
+
+
accepted NADP with about a hundred times lower efficiency than NAD ,thus
further limiting its applicability in synthetic reactions.
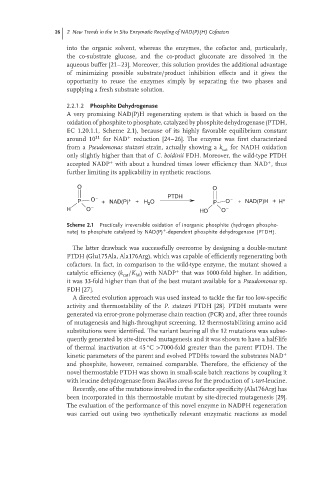
O O
P O − + NAD(P) + + H 2 O PTDH P O − + NAD(P)H + H +
H O − HO O −
Scheme 2.1 Practically irreversible oxidation of inorganic phosphite (hydrogen phospho-
+
nate) to phosphate catalyzed by NAD(P) -dependent phosphite dehydrogenase (PTDH).
The latter drawback was successfully overcome by designing a double-mutant
PTDH (Glu175Ala, Ala176Arg), which was capable of efficiently regenerating both
cofactors. In fact, in comparison to the wild-type enzyme, the mutant showed a
+
catalytic efficiency (k /K )withNADP that was 1000-fold higher. In addition,
M
cat
it was 33-fold higher than that of the best mutant available for a Pseudomonas sp.
FDH [27].
A directed evolution approach was used instead to tackle the far too low-specific
activity and thermostability of the P. stutzeri PTDH [28]. PTDH mutants were
generated via error-prone polymerase chain reaction (PCR) and, after three rounds
of mutagenesis and high-throughput screening, 12 thermostabilizing amino acid
substitutions were identified. The variant bearing all the 12 mutations was subse-
quently generated by site-directed mutagenesis and it was shown to have a half-life
◦
of thermal inactivation at 45 C >7000-fold greater than the parent PTDH. The
kinetic parameters of the parent and evolved PTDHs toward the substrates NAD +
and phosphite, however, remained comparable. Therefore, the efficiency of the
novel thermostable PTDH was shown in small-scale batch reactions by coupling it
with leucine dehydrogenase from Bacillus cereus for the production of l-tert-leucine.
Recently, one of the mutations involved in the cofactor specificity (Ala176Arg) has
been incorporated in this thermostable mutant by site-directed mutagenesis [29].
The evaluation of the performance of this novel enzyme in NADPH regeneration
was carried out using two synthetically relevant enzymatic reactions as model