Page 83 - The Biochemistry of Inorganic Polyphosphates
P. 83
March 9, 2004
15:32
Char Count= 0
WU095-06
WU095/Kulaev
Enzymes of polyphosphate biosynthesis 67
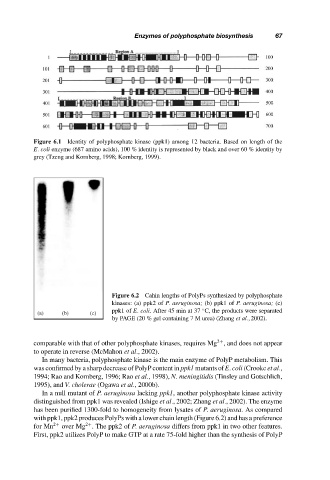
Figure 6.1 Identity of polyphosphate kinase (ppk1) among 12 bacteria. Based on length of the
E. coli enzyme (687 amino acids), 100 % identity is represented by black and over 60 % identity by
grey (Tzeng and Kornberg, 1998; Kornberg, 1999).
+
Figure 6.2 Cahin lengths of PolyPs synthesized by polyphosphate
kinases: (a) ppk2 of P. aeruginosa; (b) ppk1 of P. aeruginosa; (c)
ppk1 of E. coli. After 45 min at 37 C, the products were separated
◦
(a) (b) (c)
by PAGE (20 % gel containing 7 M urea) (Zhang et al., 2002).
comparable with that of other polyphosphate kinases, requires Mg , and does not appear
2+
to operate in reverse (McMahon et al., 2002).
In many bacteria, polyphosphate kinase is the main enzyme of PolyP metabolism. This
was confirmed by a sharp decrease of PolyP content in ppk1 mutants of E. coli (Crooke et al.,
1994; Rao and Kornberg, 1996; Rao et al., 1998), N. meningitidis (Tinsley and Gotschlich,
1995), and V. cholerae (Ogawa et al., 2000b).
In a null mutant of P. aeruginosa lacking ppk1, another polyphosphate kinase activity
distinguished from ppk1 was revealed (Ishige et al., 2002; Zhang et al., 2002). The enzyme
has been purified 1300-fold to homogeneity from lysates of P. aeruginosa. As compared
with ppk1, ppk2 produces PolyPs with a lower chain length (Figure 6.2) and has a preference
for Mn 2+ over Mg . The ppk2 of P. aeruginosa differs from ppk1 in two other features.
2+
First, ppk2 utilizes PolyP to make GTP at a rate 75-fold higher than the synthesis of PolyP