Page 169 - Algae Anatomy, Biochemistry, and Biotechnology
P. 169
152 Algae: Anatomy, Biochemistry, and Biotechnology
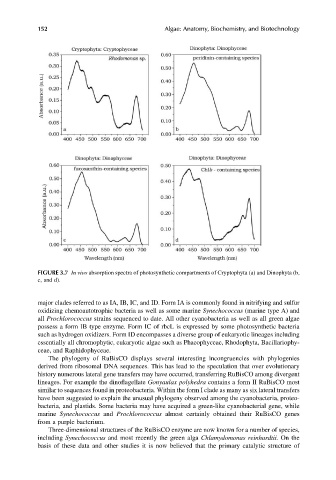
FIGURE 3.7 In vivo absorption spectra of photosynthetic compartments of Cryptophyta (a) and Dinophyta (b,
c, and d).
major clades referred to as IA, IB, IC, and ID. Form IA is commonly found in nitrifying and sulfur
oxidizing chemoautotrophic bacteria as well as some marine Synechococcus (marine type A) and
all Prochlorococcus strains sequenced to date. All other cyanobacteria as well as all green algae
possess a form IB type enzyme. Form IC of rbcL is expressed by some photosynthetic bacteria
such as hydrogen oxidizers. Form ID encompasses a diverse group of eukaryotic lineages including
essentially all chromophytic, eukaryotic algae such as Phaeophyceae, Rhodophyta, Bacillariophy-
ceae, and Raphidophyceae.
The phylogeny of RuBisCO displays several interesting incongruencies with phylogenies
derived from ribosomal DNA sequences. This has lead to the speculation that over evolutionary
history numerous lateral gene transfers may have occurred, transferring RuBisCO among divergent
lineages. For example the dinoflagellate Gonyaulax polyhedra contains a form II RuBisCO most
similar to sequences found in proteobacteria. Within the form I clade as many as six lateral transfers
have been suggested to explain the unusual phylogeny observed among the cyanobacteria, proteo-
bacteria, and plastids. Some bacteria may have acquired a green-like cyanobacterial gene, while
marine Synechococcus and Prochlorococcus almost certainly obtained their RuBisCO genes
from a purple bacterium.
Three-dimensional structures of the RuBisCO enzyme are now known for a number of species,
including Synechococcus and most recently the green alga Chlamydomonas reinhardtii. On the
basis of these data and other studies it is now believed that the primary catalytic structure of