Page 26 - Advances in Textile Biotechnology
P. 26
Design and engineering of novel enzymes for textile applications 5
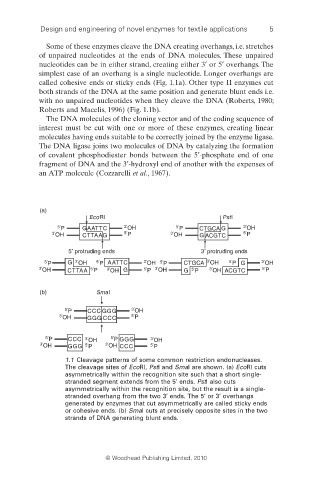
Some of these enzymes cleave the DNA creating overhangs, i.e. stretches
of unpaired nucleotides at the ends of DNA molecules. These unpaired
nucleotides can be in either strand, creating either 3′ or 5′ overhangs. The
simplest case of an overhang is a single nucleotide. Longer overhangs are
called cohesive ends or sticky ends (Fig. 1.1a). Other type II enzymes cut
both strands of the DNA at the same position and generate blunt ends i.e.
with no unpaired nucleotides when they cleave the DNA (Roberts, 1980;
Roberts and Macelis, 1996) (Fig. 1.1b).
The DNA molecules of the cloning vector and of the coding sequence of
interest must be cut with one or more of these enzymes, creating linear
molecules having ends suitable to be correctly joined by the enzyme ligase.
The DNA ligase joins two molecules of DNA by catalyzing the formation
of covalent phosphodiester bonds between the 5′-phosphate end of one
fragment of DNA and the 3′-hydroxyl end of another with the expenses of
an ATP molecule (Cozzarelli et al., 1967).
(a)
EcoRI PstI
5′ P GAATTC 3′ OH 5′ P CTGCA G 3′ OH
3′ OH CTTAAG 5′ P 3′ OH G ACGTC 5′ P
5′ protruding ends 3′ protruding ends
5′ P G 3′ OH 5′ P AATTC 3′ OH 5′ P CTGCA 3′ OH 5′ P G 3′ OH
3′ OH CTTAA 5′ P 3′ OH G 5′ P 3′ OH G 5′ P 3′ OH ACGTC 5′ P
(b) SmaI
5′ P CCC GGG 3′ OH
3′ OH GGG CCC 5′ P
5′ P CCC 3′ OH 5′ P GGG 3′ OH
3′ OH GGG 5′ P 3′ OH CCC 5′ P
1.1 Cleavage patterns of some common restriction endonucleases.
The cleavage sites of EcoRI, PstI and SmaI are shown. (a) EcoRI cuts
asymmetrically within the recognition site such that a short single-
stranded segment extends from the 5′ ends. PstI also cuts
asymmetrically within the recognition site, but the result is a single-
stranded overhang from the two 3′ ends. The 5′ or 3′ overhangs
generated by enzymes that cut asymmetrically are called sticky ends
or cohesive ends. (b) SmaI cuts at precisely opposite sites in the two
strands of DNA generating blunt ends.
© Woodhead Publishing Limited, 2010