Page 92 - Academic Press Encyclopedia of Physical Science and Technology 3rd Molecular Biology
P. 92
P1: GTQ Final pages
Encyclopedia of Physical Science and Technology EN017F-788 August 3, 2001 16:27
Translation of RNA to Protein 33
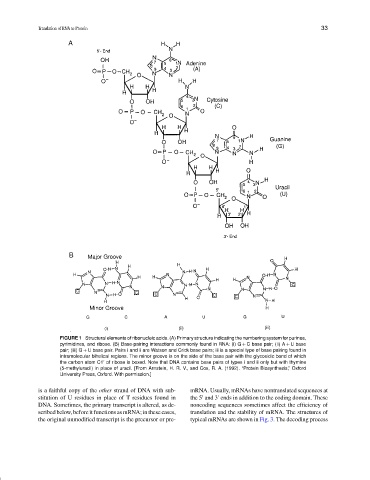
FIGURE 1 Structural elements of ribonucleic acids. (A) Primary structure indicating the numbering system for purines,
pyrimidines, and ribose. (B) Base-pairing interactions commonly found in RNA: (i) G + C base pair; (ii) A + U base
pair; (iii) G + U base pair. Pairs i and ii are Watson and Crick base pairs; iii is a special type of base pairing found in
intramolecular bihelical regions. The minor groove is on the side of the base pair with the glycosidic bond of which
the carbon atom C1 of ribose is boxed. Note that DNA contains base pairs of types i and ii only but with thymine
(5-methyluracil) in place of uracil. [From Arnstein, H. R. V., and Cox, R. A. (1992). “Protein Biosynthesis,” Oxford
University Press, Oxford. With permission.]
is a faithful copy of the other strand of DNA with sub- mRNA. Usually, mRNAs have nontranslated sequences at
stitution of U residues in place of T residues found in the 5 and 3 ends in addition to the coding domain. These
DNA. Sometimes, the primary transcript is altered, as de- noncoding sequences sometimes affect the efficiency of
scribedbelow,beforeitfunctionsasmRNA;inthesecases, translation and the stability of mRNA. The structures of
the original unmodified transcript is the precursor or pre- typical mRNAs are shown in Fig. 3. The decoding process