Page 95 - Academic Press Encyclopedia of Physical Science and Technology 3rd Molecular Biology
P. 95
P1: GTQ Final pages
Encyclopedia of Physical Science and Technology EN017F-788 August 3, 2001 16:27
36 Translation of RNA to Protein
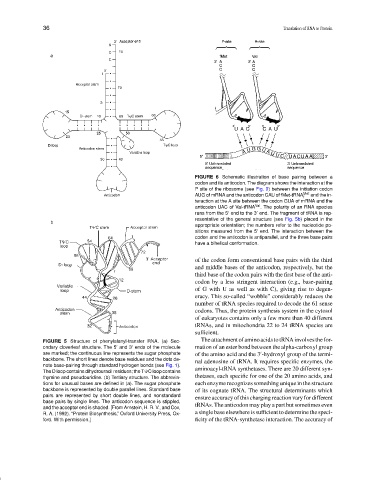
FIGURE 6 Schematic illustration of base pairing between a
codon and its anticodon. The diagram shows the interaction at the
P site of the ribosome (see Fig. 9) between the initiation codon
AUG of mRNA and the anticodon CAU of fMet-tRNA Met and the in-
f
teraction at the A site between the codon GUA of mRNA and the
anticodon UAC of Val-tRNA Val . The polarity of an RNA species
runs from the 5 end to the 3 end. The fragment of tRNA is rep-
resentative of the general structure (see Fig. 5b) placed in the
appropriate orientation; the numbers refer to the nucleotide po-
sitions measured from the 5 end. The interaction between the
codon and the anticodon is antiparallel, and the three base pairs
have a bihelical conformation.
of the codon form conventional base pairs with the third
and middle bases of the anticodon, respectively, but the
third base of the codon pairs with the first base of the anti-
codon by a less stringent interaction (e.g., base-pairing
of G with U as well as with C), giving rise to degen-
eracy. This so-called “wobble” considerably reduces the
number of tRNA species required to decode the 61 sense
codons. Thus, the protein synthesis system in the cytosol
of eukaryotes contains only a few more than 40 different
tRNAs, and in mitochondria 22 to 24 tRNA species are
sufficient.
FIGURE 5 Structure of phenylalanyl-transfer RNA. (a) Sec- TheattachmentofaminoacidstotRNAinvolvesthefor-
ondary cloverleaf structure. The 5 and 3 ends of the molecule mation of an ester bond between the alpha-carboxyl group
are marked; the continuous line represents the sugar phosphate of the amino acid and the 3 -hydroxyl group of the termi-
backbone. The short lines denote base residues and the dots de-
nal adenosine of tRNA. It requires specific enzymes, the
note base-pairing through standard hydrogen bonds (see Fig. 1).
aminoacyl-tRNA synthetases. There are 20 different syn-
The D loop contains dihydrouracil residues; the T C loop contains
thetases, each specific for one of the 20 amino acids, and
thymine and pseudouridine. (b) Tertiary structure. The abbrevia-
tions for unusual bases are defined in (a). The sugar phosphate each enzyme recognizes something unique in the structure
backbone is represented by double parallel lines. Standard base of its cognate tRNA. The structural determinants which
pairs are represented by short double lines, and nonstandard ensure accuracy of this charging reaction vary for different
base pairs by single lines. The anticodon sequence is stippled,
and the acceptor end is shaded. [From Arnstein, H. R. V., and Cox, tRNAs. The anticodon may play a part but sometimes even
R. A. (1992). “Protein Biosynthesis,” Oxford University Press, Ox- a single base elsewhere is sufficient to determine the speci-
ford. With permission.] ficity of the tRNA-synthetase interaction. The accuracy of