Page 375 - Thermodynamics of Biochemical Reactions
P. 375
Redox Reactions 375
In[lZl:= calctrGerx[ec, pHlist-, islist-] : =
Module[{energy}. (*Calculates the standard transformed Gibbs
energy of reaction in kJ mol*-1 at specified pHs and ionic
strengths for a biochemical reaction typed in the form atp+h2o+de=-
adp+pi. The names of reactants call the appropriate functions of
pH and ionic strength. pHlist and is list can be lists.*)
energy = Solve [eq, de] ;
energyul, 1, 21 /. pH + pHlist /. is + islist]
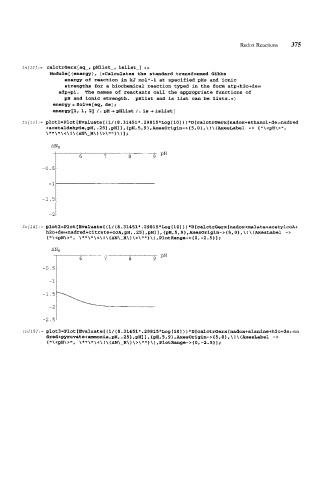
In[l3/:= glotl=Plot [Evaluate[ (1/ (8.31451*.29815*Log[lO] ) ) *D[calctrGerx[nadox+ethanol+de==nadred
+acetaldehyde,gH,.25],gH]],{pH,5,9~,AxesOrigin->{5,O},\!\(AxesLabel -> {"\<pH\>",
\*'I\ "\<\ ! \ (AN\-H\ \ >\ U" 1 \ ) I ;
)
6 7 8 9 pH
-1.5-
-2-
In[14]:= ~lot2=Plot[Evaluate[(l/(8.3l45l*.298l5*Log[lO]))*D[calctrGerx[nadox+~alate+acetylcoA+
h2o+de::nadred+citrate+coA,pH,.25],pH]],{pH,5,9},~esOrigin-~{5,O},\!\~~esLabel ->
\ * \ \ < \ ! \ ( AN\-H\ ) \ >\ 1 \ ) , Plot Range- > { 0, - 2.5 1 1 ;
{ \ <pH\ >I1,
AN,
6 7 8 9 pH
-0.51
-2.5'
In[15]:= plot3=Plot [Evaluate[ (1/ (8.31451*.29815*Log[lO] ) ) *D[calctrGerx[nadox+alanine+h2o+de==na
dred+pyruvate+ammonia,gH,.25],gH]].{gH,5,9~,AxesOrigin->(5,O~,\!\(AxesLabel ->
{ \ <pH\ > la, \ * \ \ < \ ! \ (AN\-H\ ) \ > \ '*I* 1 \ ) , PlotRange- > { 0, -2.5 1 1 ;