Page 370 - Thermodynamics of Biochemical Reactions
P. 370
370 Mathematica Solutions to Problems
calcappredpot [e~, nu-, pHlist-, islist-] : =
Module[{energy}, (*Calculates the standard apparent
reduction potential of a half reaction at specified pHs and ionic
strengths for a biochemical half reaction typed in the form
nadox+de==nadred.
The names of the reactants call the
corresponding functions of pH and ionic strength. nu is the
number of electrons involved. pHlist and islist can be lists.*)
energy = Solve [eq, de] ;
(-l*energy[[l, 1, 211 / (nu*96.485)) /. pH-tpHlist /. is+islist
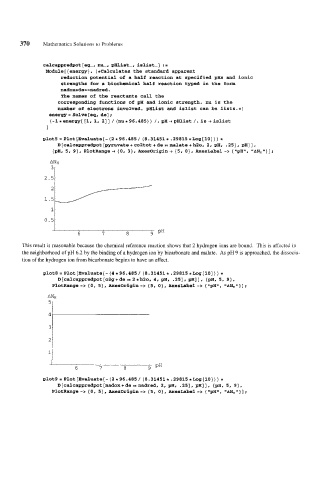
plot5 = Plot [Evaluate[- (2 * 96.485 / (8.31451 * .29815 *Log [lo] ) ) *
D [calcappredpot [pyruvate + co2tot + de == malate + h20, 2, pH, -251, pH] 1,
{BH, 5, g}, P1otRange-t {o, 3}, Axesorigin-, (5, O), AxesLabel -> {8'pH11, "ANHtl}];
I
t
6 7 8 9 pH
This result is reasonable because the chemical reference reaction shows that 2 hydrogen ions are bound. This is affected in
the neighborhood of pH 6.2 by the binding of a hydrogen ion by bicarbonate and malate. As pH 9 is approached, the dissocia-
tion of the hydrogen ion from bicarbonate begins to have an effect.
plot8 = Plot[Evaluate[- (4*96.485/ (8.31451* .29815*Log[lO])) *
D[calcappredpot [02g + de == 2 * h20, 4, pH, .25], pH] 1, {pH, 5, 9),
PlotRange -> (0, 5). Axesorigin -> {5, 0), AxesLabel -> {"pH", lrANxrm}];
AN,
A
7
3-
2.
1.
plot9 = Plot [Evaluate[- (2 * 96.485 / (8 -31451 * .29815 * Log[lO] ) ) *
D[calcappredpot[nadox+de=:nadred, 2, pH, .25], pH]], {pH, 5, 9},
PlotRange -> {0, 5}, Axesorigin -> {5, 0). AxesLabel -> {"pH", 'rANH1'}] ;