Page 141 - Advances in Textile Biotechnology
P. 141
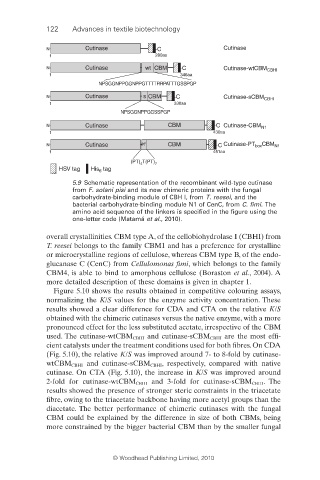
122 Advances in textile biotechnology
N Cutinase C Cutinase
1 280aa
N Cutinase wt CBM C Cutinase-wtCBM CBHI
1 346aa
NPSGGNPPGGNPPGTTTTRRPATTTGSSPGP
N Cutinase s CBM C Cutinase-sCBM CBHI
1 330aa
NPSGGNPPGGSSPGP
N Cutinase CBM C Cutinase-CBM N1
1 430aa
N Cutinase PT CBM C Cutinase-PT box CBM N1
1 451aa
(PT) T(PT) 7
4
HSV tag His tag
6
5.9 Schematic representation of the recombinant wild-type cutinase
from F. solani pisi and its new chimeric proteins with the fungal
carbohydrate-binding module of CBH I, from T. reesei, and the
bacterial carbohydrate-binding module N1 of CenC, from C. fi mi. The
amino acid sequence of the linkers is specified in the figure using the
one-letter code (Matamá et al., 2010).
overall crystallinities. CBM type A, of the cellobiohydrolase I (CBHI) from
T. reesei belongs to the family CBM1 and has a preference for crystalline
or microcrystalline regions of cellulose, whereas CBM type B, of the endo-
glucanase C (CenC) from Cellulomonas fi mi, which belongs to the family
CBM4, is able to bind to amorphous cellulose (Boraston et al., 2004). A
more detailed description of these domains is given in chapter 1.
Figure 5.10 shows the results obtained in competitive colouring assays,
normalizing the K/S values for the enzyme activity concentration. These
results showed a clear difference for CDA and CTA on the relative K/S
obtained with the chimeric cutinases versus the native enzyme, with a more
pronounced effect for the less substituted acetate, irrespective of the CBM
used. The cutinase-wtCBM CBHI and cutinase-sCBM CBHI are the most effi -
cient catalysts under the treatment conditions used for both fibres. On CDA
(Fig. 5.10), the relative K/S was improved around 7- to 8-fold by cutinase-
wtCBM CBHI and cutinase-sCBM CBHI , respectively, compared with native
cutinase. On CTA (Fig. 5.10), the increase in K/S was improved around
2-fold for cutinase-wtCBM CBHI and 3-fold for cutinase-sCBM CBHI . The
results showed the presence of stronger steric constraints in the triacetate
fi bre, owing to the triacetate backbone having more acetyl groups than the
diacetate. The better performance of chimeric cutinases with the fungal
CBM could be explained by the difference in size of both CBMs, being
more constrained by the bigger bacterial CBM than by the smaller fungal
© Woodhead Publishing Limited, 2010