Page 205 - An Introduction to Microelectromechanical Systems Engineering
P. 205
184 MEMS Applications in Life Sciences
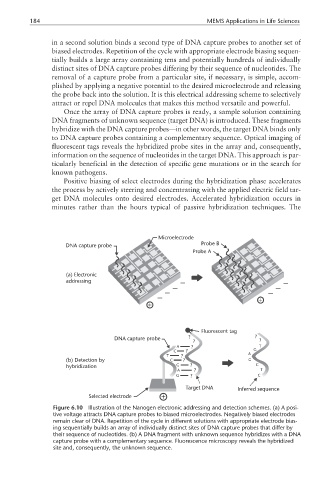
in a second solution binds a second type of DNA capture probes to another set of
biased electrodes. Repetition of the cycle with appropriate electrode biasing sequen-
tially builds a large array containing tens and potentially hundreds of individually
distinct sites of DNA capture probes differing by their sequence of nucleotides. The
removal of a capture probe from a particular site, if necessary, is simple, accom-
plished by applying a negative potential to the desired microelectrode and releasing
the probe back into the solution. It is this electrical addressing scheme to selectively
attract or repel DNA molecules that makes this method versatile and powerful.
Once the array of DNA capture probes is ready, a sample solution containing
DNA fragments of unknown sequence (target DNA) is introduced. These fragments
hybridize with the DNA capture probes—in other words, the target DNA binds only
to DNA capture probes containing a complementary sequence. Optical imaging of
fluorescent tags reveals the hybridized probe sites in the array and, consequently,
information on the sequence of nucleotides in the target DNA. This approach is par-
ticularly beneficial in the detection of specific gene mutations or in the search for
known pathogens.
Positive biasing of select electrodes during the hybridization phase accelerates
the process by actively steering and concentrating with the applied electric field tar-
get DNA molecules onto desired electrodes. Accelerated hybridization occurs in
minutes rather than the hours typical of passive hybridization techniques. The
Microelectrode
DNA capture probe Probe B
Probe A
(a) Electronic
addressing − −
− −
− −
−
Fluorescent tag
DNA capture probe ? ?
? ?
A ? T
C ? G
T ? A
(b) Detection by C ? G
hybridization G ? C
A ? T
G ? C
Target DNA Inferred sequence
Selected electrode
Figure 6.10 Illustration of the Nanogen electronic addressing and detection schemes. (a) A posi-
tive voltage attracts DNA capture probes to biased microelectrodes. Negatively biased electrodes
remain clear of DNA. Repetition of the cycle in different solutions with appropriate electrode bias-
ing sequentially builds an array of individually distinct sites of DNA capture probes that differ by
their sequence of nucleotides. (b) A DNA fragment with unknown sequence hybridizes with a DNA
capture probe with a complementary sequence. Fluorescence microscopy reveals the hybridized
site and, consequently, the unknown sequence.