Page 202 - An Introduction to Microelectromechanical Systems Engineering
P. 202
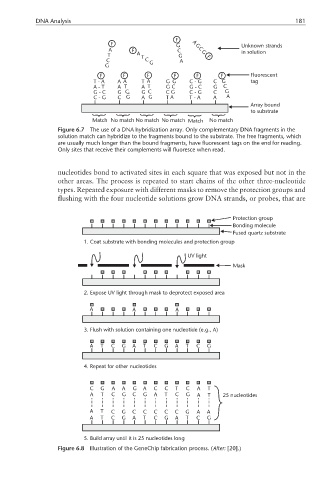
DNA Analysis 181
F
F G A G Unknown strands
A F C C
T A T G G F in solution
C C G A
G
F F F F F F Fluorescent
-
T-A A A T A G G CG C G tag
A-T A T A T G C GC G C
-
G-C G C G C C G CG C G
-
C-G C G A G T A T-A A A
Array bound
to substrate
Match No match No match No match Match No match
Figure 6.7 The use of a DNA hybridization array. Only complementary DNA fragments in the
solution match can hybridize to the fragments bound to the substrate. The free fragments, which
are usually much longer than the bound fragments, have fluorescent tags on the end for reading.
Only sites that receive their complements will fluoresce when read.
nucleotides bond to activated sites in each square that was exposed but not in the
other areas. The process is repeated to start chains of the other three-nucleotide
types. Repeated exposure with different masks to remove the protection groups and
flushing with the four nucleotide solutions grow DNA strands, or probes, that are
Protection group
Bonding molecule
Fused quartz substrate
1. Coat substrate with bonding molecules and protection group
UV light
Mask
2. Expose UV light through mask to deprotect exposed area
A A A
3. Flush with solution containing one nucleotide (e.g., A)
A T C G A T C G A T C G
4. Repeat for other nucleotides
C G A A G A C C T C A T
A T C G C G A T C G A T 25 nucleotides
A T C G C C C C C G A A
A T C G A T C G A T C G
5. Build array until it is 25 nucleotides long
Figure 6.8 Illustration of the GeneChip fabrication process. (After: [20].)