Page 210 - Applied Probability
P. 210
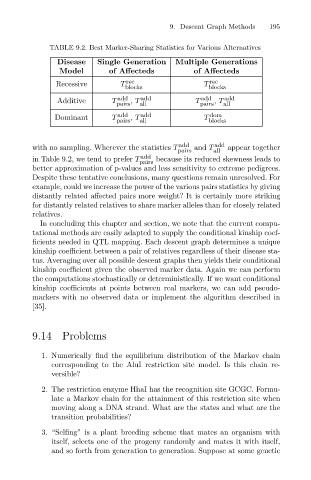
TABLE 9.2. Best Marker-Sharing Statistics for Various Alternatives
Multiple Generations
Disease
of Affecteds
Model
of Affecteds
rec
rec
T
Recessive Single Generation 9. Descent Graph Methods 195
T
blocks blocks
Additive T add , T add T add , T add
pairs all pairs all
add
dom
add
Dominant T pairs , T all T blocks
add
add
with no sampling. Wherever the statistics T pairs and T all appear together
add
in Table 9.2, we tend to prefer T pairs because its reduced skewness leads to
better approximation of p-values and less sensitivity to extreme pedigrees.
Despite these tentative conclusions, many questions remain unresolved. For
example, could we increase the power of the various pairs statistics by giving
distantly related affected pairs more weight? It is certainly more striking
for distantly related relatives to share marker alleles than for closely related
relatives.
In concluding this chapter and section, we note that the current compu-
tational methods are easily adapted to supply the conditional kinship coef-
ficients needed in QTL mapping. Each descent graph determines a unique
kinship coefficient between a pair of relatives regardless of their disease sta-
tus. Averaging over all possible descent graphs then yields their conditional
kinship coefficient given the observed marker data. Again we can perform
the computations stochastically or deterministically. If we want conditional
kinship coefficients at points between real markers, we can add pseudo-
markers with no observed data or implement the algorithm described in
[35].
9.14 Problems
1. Numerically find the equilibrium distribution of the Markov chain
corresponding to the AluI restriction site model. Is this chain re-
versible?
2. The restriction enzyme HhaI has the recognition site GCGC. Formu-
late a Markov chain for the attainment of this restriction site when
moving along a DNA strand. What are the states and what are the
transition probabilities?
3. “Selfing” is a plant breeding scheme that mates an organism with
itself, selects one of the progeny randomly and mates it with itself,
and so forth from generation to generation. Suppose at some genetic