Page 219 - Applied Probability
P. 219
10. Molecular Phylogeny
205
(2n − 1)(2n − 2)(2n − 3)!
=
2(n − 1)(n − 2)!
n−2
2
(2n − 1)!
=
.
n−1
2
(n − 1)!
For instance with n = 2, the single rooted tree of Figure 10.1 is transformed
into one of the three rooted trees of Figure 10.2 by the addition of the tip
3.
10.3 Maximum Parsimony
The first step in constructing an evolutionary tree is to partially sequence
the DNA of one representative member from each of several related taxa. A
site-by-site comparison of the bases observed along some common stretch of
DNA is then undertaken to ascertain which evolutionary tree best explains
the relationships among the taxa. In the past, evolutionary biologists have
also compared amino acid sequences deduced from one or more common
proteins. DNA sequence data are now preferred because of their greater
information content. As discussed in Appendix A, the four DNA bases are
A (adenine), G (guanine), C (cytosine), and T (thymine). Of these, two are
purines (A and G), and two are pyrimidines (C and T).
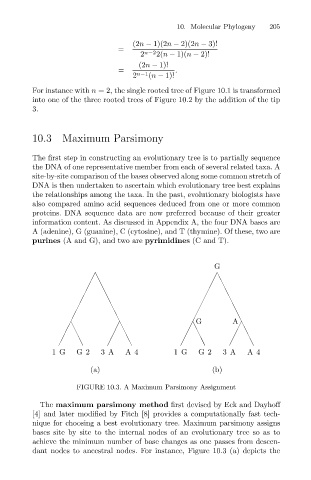
G
G A
1 G G2 3A A4 1G G2 3 A A4
(a) (b)
FIGURE 10.3. A Maximum Parsimony Assignment
The maximum parsimony method first devised by Eck and Dayhoff
[4] and later modified by Fitch [8] provides a computationally fast tech-
nique for choosing a best evolutionary tree. Maximum parsimony assigns
bases site by site to the internal nodes of an evolutionary tree so as to
achieve the minimum number of base changes as one passes from descen-
dant nodes to ancestral nodes. For instance, Figure 10.3 (a) depicts the