Page 258 - Big Data Analytics for Intelligent Healthcare Management
P. 258
10.4 DE NOVO ASSEMBLY, RE-SEQUENCING, TRANSCRIPTOMICS 251
SEQUENCING
different Kmer values. The output of short-read data, as produced by next-generation sequencing
technologies, were built quickly as long continuous sequences using the Velvet and SOAPdenovo2
assemblers. The de novo assembly was useful for the data from a new organism for which a ref-
erence sequence has not been assembled yet or to determine the origin of unmapped reads [41, 42].
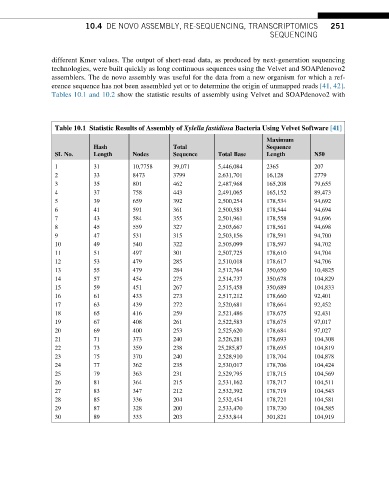
Tables 10.1 and 10.2 show the statistic results of assembly using Velvet and SOAPdenovo2 with
Table 10.1 Statistic Results of Assembly of Xylella fastidiosa Bacteria Using Velvet Software [41]
Maximum
Hash Total Sequence
SL No. Length Nodes Sequence Total Base Length N50
1 31 10,7758 39,071 5,446,084 2365 207
2 33 8473 3799 2,631,701 16,128 2779
3 35 801 462 2,487,968 165,208 79,655
4 37 758 443 2,491,065 165,152 89,473
5 39 659 392 2,500,254 178,534 94,692
6 41 591 361 2,500,583 178,544 94,694
7 43 584 355 2,501,961 178,558 94,696
8 45 559 327 2,503,667 178,561 94,698
9 47 531 315 2,503,156 178,591 94,700
10 49 540 322 2,505,099 178,597 94,702
11 51 497 301 2,507,725 178,610 94,704
12 53 479 285 2,510,018 178,617 94,706
13 55 479 284 2,512,764 350,650 10,4825
14 57 454 275 2,514,737 350,678 104,829
15 59 451 267 2,515,458 350,689 104,833
16 61 433 273 2,517,212 178,660 92,401
17 63 439 272 2,520,681 178,664 92,452
18 65 416 259 2,521,486 178,675 92,431
19 67 408 261 2,522,583 178,675 97,017
20 69 400 253 2,525,620 178,684 97,027
21 71 373 240 2,526,281 178,693 104,308
22 73 359 238 25,285,87 178,695 104,819
23 75 370 240 2,528,910 178,704 104,878
24 77 362 235 2,530,017 178,706 104,424
25 79 363 231 2,529,795 178,715 104,569
26 81 364 215 2,531,162 178,717 104,511
27 83 347 212 2,532,392 178,719 104,543
28 85 336 204 2,532,454 178,721 104,581
29 87 328 200 2,533,470 178,730 104,585
30 89 333 203 2,533,844 301,821 104,919