Page 297 - Cascade_Biocatalysis_Integrating_Stereoselective_and_Environmentally_Friendly_Reactions
P. 297
12.2 Diversity of Nitrilase Sequences 273
Table 12.1 Characterized nitrilase and cyanide hydratases predicted from sequence data.
Enzyme Strain Sequence ID References
Cyanide hydratase Aspergillus nidulans FGSC #A4 tpe|CBF78050.1| a [5]
Neurospora crassa FGSC #2489 ref|XP 960160.2|
Gibberella zeae ref|XP 385981.1|
Aspergillus niger CCF 3411 gb|ABX75546.1| [6]
Penicillium chrysogenum ref|XP 002562104.1|
Wisconsin 54-1255
Arylacetonitrilase Neurospora crassa OR74A emb|CAD70472.1| [6, 7]
Aspergillus niger CBS 513.88 ref|XP 001397369.1|
[6]
ref|XP 001398633.1|
Aspergillus oryzae RIB40 ref|XP 001824712.1|
Arthroderma benhamiae CBS ref|XP 003011330.1| [8, 6]
112371
Nectria haematococca mpVI ref|XP 003050920.1|
77-13-4
Burkholderia xenovorans LB400 ref|YP 559838.1| [9]
Bradyrhizobium japonicum ref|NP 773042.1| [9, 10]
USDA110
[11]
ref|NP 770037.1|
Labrenzia aggregata DSM gb|EAV41725.1| [30]
13394
Aromatic nitrilase Gibberella moniliformis gb|ABF83489.1| [6, 7]
Penicillium marneffei ref|XP 002144951.1|
ATCC18224
Penicillium chrysogenum ref|XP 002565836.1| [6]
Wisconsin 54-1255
Meyerozyma guilliermondi ATCC ref|XP 001482890.1|
6260
Aliphatic nitrilases Synechocystis sp. PCC6803 ref|YP 005384515.1| [12]
Pyrococcus abyssi GE5 pdb|3IVZ|A [13, 14]
a The protein expressed in E. coli differed in length from the hypothetical protein in the same strain
(356 vs 364 amino acids). The authors assumed that the introns were not correctly determined in the
corresponding gene [5].
H 2 O
NH 3
E-SH S E
RC N H 2 O RC O RC OH
SE
SE O
RC NH 2 E-SH
RC NH
OH
RC NH 2
E-SH O
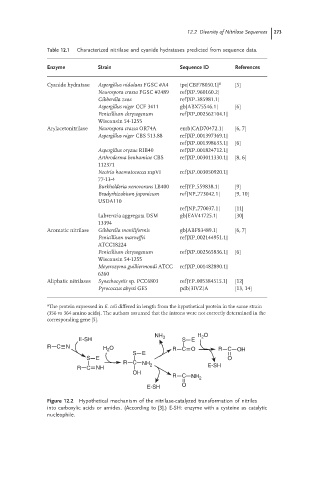
Figure 12.2 Hypothetical mechanism of the nitrilase-catalyzed transformation of nitriles
into carboxylic acids or amides. (According to [3].) E-SH: enzyme with a cysteine as catalytic
nucleophile.