Page 327 - Biosystems Engineering
P. 327
304 Cha pte r Ni ne
in the affinity chromatographic process. A good matrix for this method
should have the following properties:
1. Hydrophilic: Reduce the nonspecific interactions.
2. Large pores: Allow all areas of the matrix to be available to
most of the molecules in the mixture. Some matrices allow
binding only to the outer surface and some are useful for
separating very large molecules, cells, or viruses.
3. Rigid: The matrix must withstand the pressures of packing
and solvent flow during elution or washing.
4. Inert: The matrix should not contribute to the separation.
5. Chemical stability: The matrix must be stable to all solvents in
the separation.
The ligand is covalently attached to supporting medium so that
the chromatographic material can be designed for a specific purifica-
tion task. A spacer arm is used between the matrix and the ligand so
that the active site of the ligand is available to the sample. Generally,
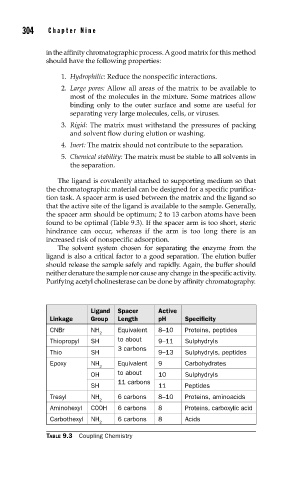
the spacer arm should be optimum; 2 to 13 carbon atoms have been
found to be optimal (Table 9.3). If the spacer arm is too short, steric
hindrance can occur, whereas if the arm is too long there is an
increased risk of nonspecific adsorption.
The solvent system chosen for separating the enzyme from the
ligand is also a critical factor to a good separation. The elution buffer
should release the sample safely and rapidly. Again, the buffer should
neither denature the sample nor cause any change in the specific activity.
Purifying acetyl cholinesterase can be done by affinity chromatography.
Ligand Spacer Active
Linkage Group Length pH Specificity
CNBr NH Equivalent 8–10 Proteins, peptides
2
Thiopropyl SH to about 9–11 Sulphydryls
3 carbons
Thio SH 9–13 Sulphydryls, peptides
Epoxy NH Equivalent 9 Carbohydrates
2
OH to about 10 Sulphydryls
11 carbons
SH 11 Peptides
Tresyl NH 6 carbons 8–10 Proteins, aminoacids
2
Aminohexyl C00H 6 carbons 8 Proteins, carboxylic acid
Carbothexyl NH 6 carbons 8 Acids
2
TABLE 9.3 Coupling Chemistry