Page 382 - Carrahers_Polymer_Chemistry,_Eighth_Edition
P. 382
Naturally Occurring Polymers—Animals 345
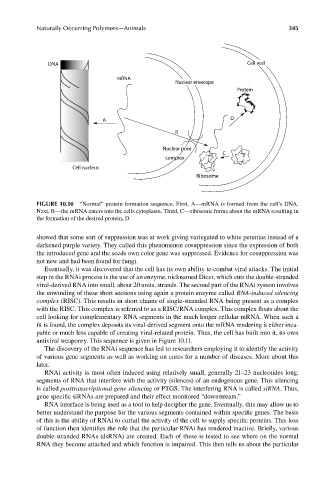
DNA Cell wall
mRNA
Nuclear envelope
Protein
A D
B
Nuclear pore
C
complex
Cell nucleus
Ribosome
FIGURE 10.10 “Normal” protein formation sequence. First, A—mRNA is formed from the cell’s DNA.
Next, B—the mRNA enters into the cells cytoplasm. Third, C—ribosome forms about the mRNA resulting in
the formation of the desired protein, D.
showed that some sort of suppression was at work giving variegated to white petunias instead of a
darkened purple variety. They called this phenomenon cosuppression since the expression of both
the introduced gene and the seeds own color gene was suppressed. Evidence for cosuppression was
not new and had been found for fungi.
Eventually, it was discovered that the cell has its own ability to combat viral attacks. The initial
step in the RNAi process is the use of an enzyme, nicknamed Dicer, which cuts the double-stranded
viral-derived RNA into small, about 20 units, strands. The second part of the RNAi system involves
the unwinding of these short sections using again a protein enzyme called RNA-induced silencing
complex (RISC). This results in short chains of single-stranded RNA being present as a complex
with the RISC. This complex is referred to as a RISC/RNA complex. This complex floats about the
cell looking for complementary RNA segments in the much longer cellular mRNA. When such a
fit is found, the complex deposits its viral-derived segment onto the mRNA rendering it either inca-
pable or much less capable of creating viral-related protein. Thus, the cell has built into it, its own
antiviral weaponry. This sequence is given in Figure 10.11.
The discovery of the RNAi sequence has led to researchers employing it to identify the activity
of various gene segments as well as working on cures for a number of diseases. More about this
later.
RNAi activity is most often induced using relatively small, generally 21–23 nucleotides long,
segments of RNA that interfere with the activity (silences) of an endogenous gene. This silencing
is called posttranscriptional gene silencing or PTGS. The interfering RNA is called siRNA. Thus,
gene specific siRNAs are prepared and their effect monitored “downstream.”
RNA interface is being used as a tool to help decipher the gene. Eventually, this may allow us to
better understand the purpose for the various segments contained within specific genes. The basis
of this is the ability of RNAi to curtail the activity of the cell to supply specifi c proteins. This loss
of function then identifies the role that the particular RNAi has rendered inactive. Briefl y, various
double-stranded RNAs (dsRNA) are created. Each of these is tested to see where on the normal
RNA they become attached and which function is impaired. This then tells us about the particular
9/14/2010 3:41:17 PM
K10478.indb 345 9/14/2010 3:41:17 PM
K10478.indb 345