Page 109 - Academic Press Encyclopedia of Physical Science and Technology 3rd BioTechnology
P. 109
P1: GNB Final Pages
Encyclopedia of Physical Science and Technology EN005F-954 June 15, 2001 20:48
822 Fiber-Optic Chemical Sensors
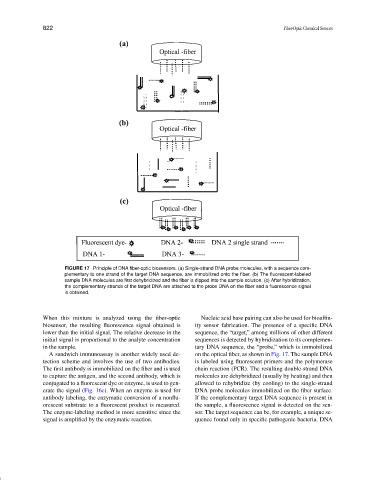
FIGURE 17 Principle of DNA fiber-optic biosensors. (a) Single-strand DNA probe molecules, with a sequence com-
plementary to one strand of the target DNA sequence, are immobilized onto the fiber. (b) The fluorescent-labeled
sample DNA molecules are first dehybridized and the fiber is dipped into the sample solution. (c) After hybridization,
the complementary strands of the target DNA are attached to the probe DNA on the fiber and a fluorescence signal
is obtained.
When this mixture is analyzed using the fiber-optic Nucleic acid base pairing can also be used for bioaffin-
biosensor, the resulting fluorescence signal obtained is ity sensor fabrication. The presence of a specificDNA
lower than the initial signal. The relative decrease in the sequence, the “target,” among millions of other different
initial signal is proportional to the analyte concentration sequences is detected by hybridization to its complemen-
in the sample. tary DNA sequence, the “probe,” which is immobilized
A sandwich immunoassay is another widely used de- on the optical fiber, as shown in Fig. 17. The sample DNA
tection scheme and involves the use of two antibodies. is labeled using fluorescent primers and the polymerase
The first antibody is immobilized on the fiber and is used chain reaction (PCR). The resulting double-strand DNA
to capture the antigen, and the second antibody, which is molecules are dehybridized (usually by heating) and then
conjugated to a fluorescent dye or enzyme, is used to gen- allowed to rehybridize (by cooling) to the single-strand
erate the signal (Fig. 16c). When an enzyme is used for DNA probe molecules immobilized on the fiber surface.
antibody labeling, the enzymatic conversion of a nonflu- If the complementary target DNA sequence is present in
orescent substrate to a fluorescent product is measured. the sample, a fluorescence signal is detected on the sen-
The enzyme-labeling method is more sensitive since the sor. The target sequence can be, for example, a unique se-
signal is amplified by the enzymatic reaction. quence found only in specific pathogenic bacteria. DNA