Page 35 - Academic Press Encyclopedia of Physical Science and Technology 3rd Polymer
P. 35
P1: FPP 2nd Revised Pages
Encyclopedia of Physical Science and Technology EN002C-64 May 19, 2001 20:39
Biopolymers 239
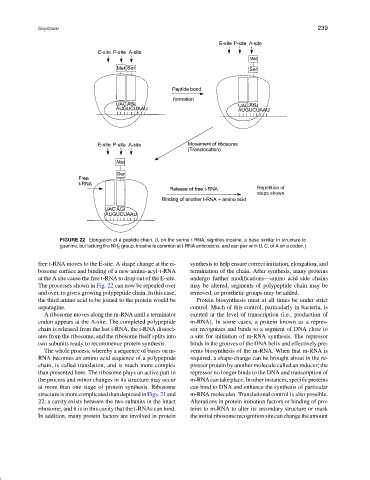
FIGURE 22 Elongation of a peptide chain. (I, on the serine t-RNA, signifies inosine, a base similar in structure to
guanine, but lacking the NH 2 group. Inosine is common at t-RNA anticodons, and can pair with U, C, or A on a codon.)
free t-RNA moves to the E-site. A shape change at the ri- synthesis to help ensure correct initiation, elongation, and
bosome surface and binding of a new amino-acyl-t-RNA termination of the chain. After synthesis, many proteins
at the A site cause the free t-RNA to drop out of the E-site. undergo further modifications—amino acid side chains
The processes shown in Fig. 22 can now be repeated over may be altered, segments of polypeptide chain may be
and over, to give a growing polypeptide chain. In this case, removed, or prosthetic groups may be added.
the third amino acid to be joined to the protein would be Protein biosynthesis must at all times be under strict
asparagine. control. Much of this control, particularly in bacteria, is
A ribosome moves along the m-RNA until a terminator exerted at the level of transcription (i.e., production of
codon appears at the A-site. The completed polypeptide m-RNA). In some cases, a protein known as a repres-
chain is released from the last t-RNA, the t-RNA dissoci- sor recognizes and binds to a segment of DNA close to
ates from the ribosome, and the ribosome itself splits into a site for initiation of m-RNA synthesis. The repressor
two subunits ready to recommence protein synthesis. binds in the grooves of the DNA helix and effectively pre-
The whole process, whereby a sequence of bases on m- vents biosynthesis of the m-RNA. When that m-RNA is
RNA becomes an amino acid sequence of a polypeptide required, a shape-change can be brought about in the re-
chain, is called translation, and is much more complex pressor protein by another molecule called an inducer; the
than presented here. The ribosome plays an active part in repressor no longer binds to the DNA and transcription of
the process and minor changes in its structure may occur m-RNAcantakeplace.Inotherinstances,specificproteins
at more than one stage of protein synthesis. Ribosome can bind to DNA and enhance the synthesis of particular
structure is more complicated than depicted in Figs. 21 and m-RNA molecules. Translational control is also possible.
22; a cavity exists between the two subunits in the intact Alterations in protein initiation factors or binding of pro-
ribosome, and it is in this cavity that the t-RNAs can bind. teins to m-RNA to alter its secondary structure or mask
In addition, many protein factors are involved in protein theinitialribosomerecognitionsitecanchangetheamount