Page 27 - Academic Press Encyclopedia of Physical Science and Technology 3rd Molecular Biology
P. 27
P1: GNH Revised Pages
Encyclopedia of Physical Science and Technology EN002G-104 May 17, 2001 20:53
Chromatin Structure and Modification 819
underlying DNA like a maze of positively charged tenta-
cles. Thus, the alterations in the charge of the tails effected
by postranslational covalent modifications are likely to
have impact on chromatin.
E. “... Not by Beads Alone”: Higher-Order
Chromatin Structure
It is clear that the nucleosome as a tool is not sufficient to
compact the entirety of the genome into the nucleus; quite
a feat of condensing is required to convert the “beads-on-
a-string”fiber into the dramatic metaphase chromosomes
so familiar to many (if the entirety of the genome were
assembled into a fully extended nucleosomal array, its
length would be ca. 15 cm).
One important functional component of further folding
by the nucleosomal fiber is the linker histone (H1). As FIGURE 10 Model for higher-order chromatin folding.
its name implies, its binding site is with the DNA stretch
between two adjacent octamer particles; the precise man-
ner in which the linker histone binds to that DNA has cording to which the nucleosomal fiber winds onto itself
been the subject of much investigation and controversy to form a solenoidal structure, with ca. six nucleosomes
(Fig. 9). Whatever the precise mode of its association per turn of the superhelix. Remarkably, whether such an
with DNA, one important consequence of this association entity forms in vivo, and what the precise arrangement is
is charge neutralization over the linker DNA stretch—that of the nucleosome within this structure, remains an open
is, the shielding of the negatively charged phosphate back- issue, and other models for this fiber have been proposed
bone from solution. Experiments in the laboratory of M. by C. Woodcock and several other scientists.
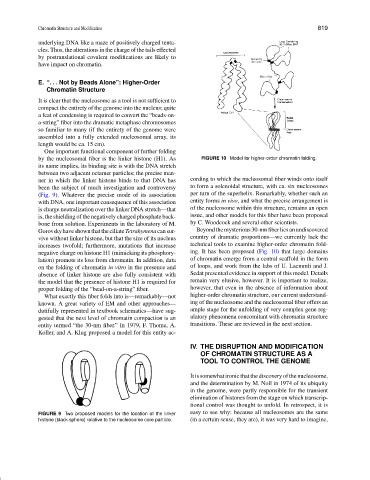
Gorovskyhaveshownthattheciliate Tetrahymenacansur- Beyondthemysterious30-nmfiberliesanundiscovered
vive without linker histone, but that the size of its nucleus country of dramatic proportions—we currently lack the
increases twofold; furthermore, mutations that increase technical tools to examine higher-order chromatin fold-
negative charge on histone H1 (mimicking its phosphory- ing. It has been proposed (Fig. 10) that large domains
lation) promote its loss from chromatin. In addition, data of chromatin emerge from a central scaffold in the form
on the folding of chromatin in vitro in the presence and of loops, and work from the labs of U. Laemmli and J.
absence of linker histone are also fully consistent with Sedat presented evidence in support of this model. Details
the model that the presence of histone H1 is required for remain very elusive, however. It is important to realize,
proper folding of the “bead-on-a-string”fiber. however, that even in the absence of information about
What exactly this fiber folds into is—remarkably—not higher-order chromatin structure, our current understand-
known. A great variety of EM and other approaches— ing of the nucleosome and the nucleosomal fiber offers an
dutifully represented in textbook schematics—have sug- ample stage for the unfolding of very complex gene reg-
gested that the next level of chromatin compaction is an ulatory phenomena concomitant with chromatin structure
entity termed “the 30-nm fiber.” In 1979, F. Thoma, A. transitions. These are reviewed in the next section.
Koller, and A. Klug proposed a model for this entity ac-
IV. THE DISRUPTION AND MODIFICATION
OF CHROMATIN STRUCTURE AS A
TOOL TO CONTROL THE GENOME
It is somewhat ironic that the discovery of the nucleosome,
and the determination by M. Noll in 1974 of its ubiquity
in the genome, were partly responsible for the transient
elimination of histones from the stage on which transcrip-
tional control was thought to unfold. In retrospect, it is
FIGURE 9 Two proposed models for the location of the linker easy to see why: because all nucleosomes are the same
histone (black sphere) relative to the nucleosome core particle. (in a certain sense, they are), it was very hard to imagine,