Page 24 - Academic Press Encyclopedia of Physical Science and Technology 3rd Molecular Biology
P. 24
P1: GNH Revised Pages
Encyclopedia of Physical Science and Technology EN002G-104 May 17, 2001 20:53
816 Chromatin Structure and Modification
by the maze of barrels and ribbons in diagrams—is to real-
ize that the globular domain of all core histones assumes
an identical secondary and tertiary structure (Fig. 7). In
this “histone fold” motif, the central, extended α helix is
flanked on each end by a short turn (loop) of the polypep-
tide, and then—again, on each end—a shorter α helix (i.e.,
somewhat like the letter “Z”). The shorter helices reside
on top of the same side of the longer helix—at its opposite
ends, of course—and both lie at an approximately right an-
gle to it. An identity of folding into this motif allows two
histone molecules to heterodimerize via a “handshake”:
the central α helices of histones H3 and H4 fit together
diagonally to form an “X” (extending the “handshake”
analogy, this is equivalent to the touching of palms), and
the shorter helices on each side of the central helix project
toward their counterparts on the other histone molecule
(analogous to fingers of one hand wrapping around the
other hand). Histones H2A and H2B interact via an iden-
tical handshake.
In terms of binding DNA, the most immediate result of
two histones coming together in a structure like this one is
that the interhelical turns (loops) of each histone molecule
become juxtaposed on each end (“top” and “bottom”)of
the “X” that is formed. These loops build a ramp onto
which DNA is wound (each histone dimer associates with
ca. 2.5 turns of DNA, i.e., ca. 27–28 base pairs); the shorter
helicesononeendofeachhistoneprojecttowardthe“front
end” of the “X” and also contact the DNA.
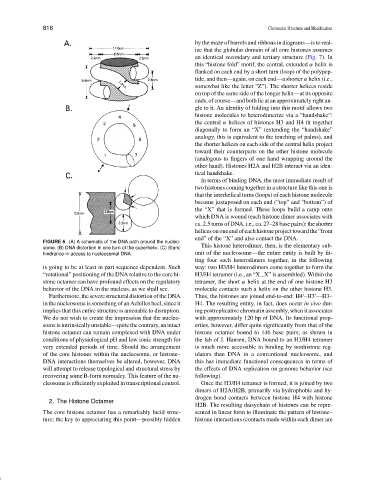
FIGURE 6 (A) A schematic of the DNA path around the nucleo-
This histone heterodimer, then, is the elementary sub-
some. (B) DNA distortion in one turn of the superhelix. (C) Steric
hindrance in access to nucleosomal DNA. unit of the nucleosome—the entire entity is built by fit-
ting four such heterodimers together, in the following
is going to be at least in part sequence dependent. Such way: two H3/H4 heterodimers come together to form the
“rotational” positioning of the DNA relative to the core hi- H3/H4 tetramer (i.e., an “X X” is assembled). Within the
stone octamer can have profound effects on the regulatory tetramer, the short α helix at the end of one histone H3
behavior of the DNA in the nucleus, as we shall see. molecule contacts such a helix on the other histone H3.
Furthermore,theseverestructuraldistortionoftheDNA Thus, the histones are joined end-to-end: H4 –H3 —H3–
in the nucleosome is something of an Achilles heel, since it H4. The resulting entity, in fact, does occur in vivo dur-
implies that this entire structure is amenable to disruption. ing postreplicative chromatin assembly, when it associates
We do not wish to create the impression that the nucleo- with approximately 120 bp of DNA. Its functional prop-
someisintrinsicallyunstable—quitethecontrary,anintact erties, however, differ quite significantly from that of the
histone octamer can remain complexed with DNA under histone octamer bound to 146 base puirs; as shown in
conditions of physiological pH and low ionic strength for the lab of J. Hansen, DNA bound to an H3/H4 tetramer
very extended periods of time. Should the arrangement is much more accessible to binding by nonhistone reg-
of the core histones within the nucleosome, or histone– ulators than DNA in a conventional nucleosome, and
DNA interactions themselves be altered, however, DNA this has immediate functional consequences in terms of
will attempt to release topological and structural stress by the effects of DNA replication on genome behavior (see
recovering some B-form normalcy. This feature of the nu- following).
cleosome is efficiently exploited in transcriptional control. Once the H3/H4 tetramer is formed, it is joined by two
dimers of H2A/H2B, primarily via hydrophobic and hy-
drogen bond contacts between histone H4 with histone
2. The Histone Octamer
H2B. The resulting daisychain of histones can be repre-
The core histone octamer has a remarkably lucid struc- sented in linear form to illuminate the pattern of histone–
ture; the key to appreciating this point—possibly hidden histone interactions (contacts made within each dimer are