Page 86 - Multidimensional Chromatography
P. 86
78 Multidimensional Chromatography
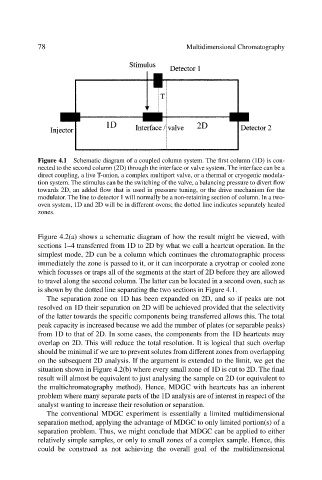
Figure 4.1 Schematic diagram of a coupled column system. The first column (1D) is con-
nected to the second column (2D) through the interface or valve system. The interface can be a
direct coupling, a live T-union, a complex multiport valve, or a thermal or cryogenic modula-
tion system. The stimulus can be the switching of the valve, a balancing pressure to divert flow
towards 2D, an added flow that is used in pressure tuning, or the drive mechanism for the
modulator. The line to detector 1 will normally be a non-retaining section of column. In a two-
oven system, 1D and 2D will be in different ovens; the dotted line indicates separately heated
zones.
Figure 4.2(a) shows a schematic diagram of how the result might be viewed, with
sections 1–4 transferred from 1D to 2D by what we call a heartcut operation. In the
simplest mode, 2D can be a column which continues the chromatographic process
immediately the zone is passed to it, or it can incorporate a cryotrap or cooled zone
which focusses or traps all of the segments at the start of 2D before they are allowed
to travel along the second column. The latter can be located in a second oven, such as
is shown by the dotted line separating the two sections in Figure 4.1.
The separation zone on 1D has been expanded on 2D, and so if peaks are not
resolved on 1D their separation on 2D will be achieved provided that the selectivity
of the latter towards the specific components being transferred allows this. The total
peak capacity is increased because we add the number of plates (or separable peaks)
from 1D to that of 2D. In some cases, the components from the 1D heartcuts may
overlap on 2D. This will reduce the total resolution. It is logical that such overlap
should be minimal if we are to prevent solutes from different zones from overlapping
on the subsequent 2D analysis. If the argument is extended to the limit, we get the
situation shown in Figure 4.2(b) where every small zone of 1D is cut to 2D. The final
result will almost be equivalent to just analysing the sample on 2D (or equivalent to
the multichromatography method). Hence, MDGC with heartcuts has an inherent
problem where many separate parts of the 1D analysis are of interest in respect of the
analyst wanting to increase their resolution or separation.
The conventional MDGC experiment is essentially a limited multidimensional
separation method, applying the advantage of MDGC to only limited portion(s) of a
separation problem. Thus, we might conclude that MDGC can be applied to either
relatively simple samples, or only to small zones of a complex sample. Hence, this
could be construed as not achieving the overall goal of the multidimensional