Page 125 - Nanotechnology an introduction
P. 125
bonds, but at a slower rate than the reaction being catalyzed.
3. The product–enzyme complex is decomposed, i.e., the products are released: AẼB → A + B + Ẽ*. Release of the products from the active
site again creates strain between it and the rest of the protein molecule.
4. Finally, the strained enzyme slowly relaxes back to its initial conformation: Ẽ* → E.
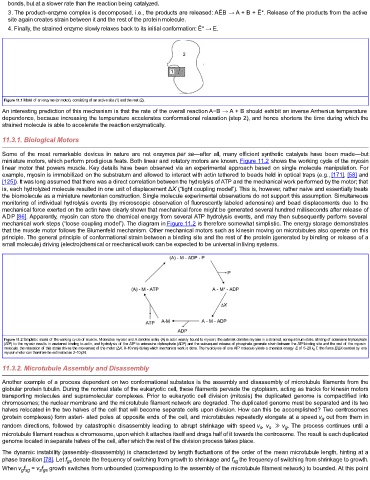
Figure 11.1 Model of an enzyme (or motor), consisting of an active site (1) and the rest (2).
An interesting prediction of this mechanism is that the rate of the overall reaction A−B → A + B should exhibit an inverse Arrhenius temperature
dependence, because increasing the temperature accelerates conformational relaxation (step 2), and hence shortens the time during which the
strained molecule is able to accelerate the reaction enzymatically.
11.3.1. Biological Motors
Some of the most remarkable devices in nature are not enzymes per se—after all, many efficient synthetic catalysts have been made—but
miniature motors, which perform prodigious feats. Both linear and rotatory motors are known. Figure 11.2 shows the working cycle of the myosin
linear motor that powers muscle. Key details have been observed via an experimental approach based on single molecule manipulation. For
example, myosin is immobilized on the substratum and allowed to interact with actin tethered to beads held in optical traps (e.g., [171], [58] and
[125]). It was long assumed that there was a direct correlation between the hydrolysis of ATP and the mechanical work performed by the motor; that
is, each hydrolyzed molecule resulted in one unit of displacement ΔX (“tight coupling model”). This is, however, rather naive and essentially treats
the biomolecule as a miniature newtonian construction. Single molecule experimental observations do not support this assumption. Simultaneous
monitoring of individual hydrolysis events (by microscopic observation of fluorescently labeled adenosine) and bead displacements due to the
mechanical force exerted on the actin have clearly shown that mechanical force might be generated several hundred milliseconds after release of
ADP [86]. Apparently, myosin can store the chemical energy from several ATP hydrolysis events, and may then subsequently perform several
mechanical work steps (“loose coupling model”). The diagram in Figure 11.2 is therefore somewhat simplistic. The energy storage demonstrates
that the muscle motor follows the Blumenfeld mechanism. Other mechanical motors such as kinesin moving on microtubules also operate on this
principle. The general principle of conformational strain between a binding site and the rest of the protein (generated by binding or release of a
small molecule) driving (electro)chemical or mechanical work can be expected to be universal in living systems.
Figure 11.2 Simplistic model of the working cycle of muscle. M denotes myosin and A denotes actin; (A) is actin weakly bound to myosin; the asterisk denotes myosin in a strained, nonequilibrium state. Binding of adenosine triphosphate
(ATP) to the myosin results in weakened binding to actin, and hydrolysis of the ATP to adenosine diphosphate (ADP) and the subsequent release of phosphate generate strain between the ATP-binding site and the rest of the myosin
molecule; the relaxation of this strain drives the movement of the motor (ΔX, 8–10 nm) during which mechanical work is done. The hydrolysis of one ATP molecule yields a chemical energy E of 5–20 k B T; the force E/ΔX exerted by one
myosin motor can therefore be estimated as 2–10 pN.
11.3.2. Microtubule Assembly and Disassembly
Another example of a process dependent on two conformational substates is the assembly and disassembly of microtubule filaments from the
globular protein tubulin. During the normal state of the eukaryotic cell, these filaments pervade the cytoplasm, acting as tracks for kinesin motors
transporting molecules and supramolecular complexes. Prior to eukaryotic cell division (mitosis) the duplicated genome is compactified into
chromosomes; the nuclear membrane and the microtubule filament network are degraded. The duplicated genome must be separated and its two
halves relocated in the two halves of the cell that will become separate cells upon division. How can this be accomplished? Two centrosomes
(protein complexes) form asteri- ated poles at opposite ends of the cell, and microtubules repeatedly elongate at a speed v out from them in
g
random directions, followed by catastrophic disassembly leading to abrupt shrinkage with speed v , v ≫ v . The process continues until a
s
g
s
microtubule filament reaches a chromosome, upon which it attaches itself and drags half of it towards the centrosome. The result is each duplicated
genome located in separate halves of the cell, after which the rest of the division process takes place.
The dynamic instability (assembly–disassembly) is characterized by length fluctuations of the order of the mean microtubule length, hinting at a
phase transition [78]. Let f denote the frequency of switching from growth to shrinkage and f the frequency of switching from shrinkage to growth.
sg
gs
When v f = v f growth switches from unbounded (corresponding to the assembly of the microtubule filament network) to bounded. At this point
g sg
s gs