Page 130 - The Biochemistry of Inorganic Polyphosphates
P. 130
WU095/Kulaev
WU095-07
Functions of polyphosphate and polyphosphate-dependent enzymes
114 March 9, 2004 15:39 Char Count= 0
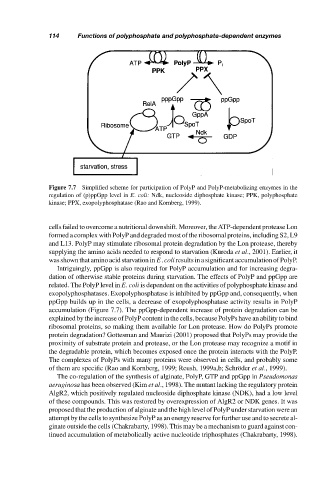
Figure 7.7 Simplified scheme for participation of PolyP and PolyP-metabolizing enzymes in the
regulation of (p)ppGpp level in E. coli: Ndk, nucleoside diphosphate kinase; PPK, polyphosphate
kinase; PPX, exopolyphosphatase (Rao and Kornberg, 1999).
cells failed to overcome a nutritional downshift. Moreover, the ATP-dependent protease Lon
formed a complex with PolyP and degraded most of the ribosomal proteins, including S2, L9
and L13. PolyP may stimulate ribosomal protein degradation by the Lon protease, thereby
supplying the amino acids needed to respond to starvation (Kuroda et al., 2001). Earlier, it
wasshownthataminoacidstarvationin E.coliresultsinasignificantaccumulationofPolyP.
Intriguingly, ppGpp is also required for PolyP accumulation and for increasing degra-
dation of otherwise stable proteins during starvation. The effects of PolyP and ppGpp are
related. The PolyP level in E. coli is dependent on the activities of polyphosphate kinase and
exopolyphosphatases. Exopolyphosphatase is inhibited by ppGpp and, consequently, when
ppGpp builds up in the cells, a decrease of exopolyphosphatase activity results in PolyP
accumulation (Figure 7.7). The ppGpp-dependent increase of protein degradation can be
explained by the increase of PolyP content in the cells, because PolyPs have an ability to bind
ribosomal proteins, so making them available for Lon protease. How do PolyPs promote
protein degradation? Gottesman and Maurizi (2001) proposed that PolyPs may provide the
proximity of substrate protein and protease, or the Lon protease may recognize a motif in
the degradable protein, which becomes exposed once the protein interacts with the PolyP.
The complexes of PolyPs with many proteins were observed in cells, and probably some
of them are specific (Rao and Kornberg, 1999; Reush, 1999a,b; Schr¨oder et al., 1999).
The co-regulation of the synthesis of alginate, PolyP, GTP and ppGpp in Pseudomonas
aeruginosa has been observed (Kim et al., 1998). The mutant lacking the regulatory protein
AlgR2, which positively regulated nucleoside diphosphate kinase (NDK), had a low level
of these compounds. This was restored by overexpression of AlgR2 or NDK genes. It was
proposed that the production of alginate and the high level of PolyP under starvation were an
attempt by the cells to synthesize PolyP as an energy reserve for further use and to secrete al-
ginate outside the cells (Chakrabarty, 1998). This may be a mechanism to guard against con-
tinued accumulation of metabolically active nucleotide triphosphates (Chakrabarty, 1998).