Page 132 - The Biochemistry of Inorganic Polyphosphates
P. 132
WU095/Kulaev
WU095-07
Functions of polyphosphate and polyphosphate-dependent enzymes
116 March 9, 2004 15:39 Char Count= 0
Lon
proteinase
RelA SpoT
Ribosome
proteins
Stationary growth
exopolyPases phase adaptation
Exopolyp (p)ppGpp
Quorum sensing
ppk
PolyP rpoS
Adhesion and
biofilm formation
virulence
NTP
Motility
RNA
degradosome
Heat, high salinity,
H O , UV light,
2 2
antiserum stability
SOS genes,
independent of
rpoS
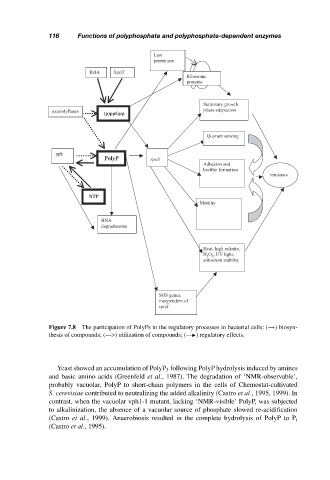
Figure 7.8 The participation of PolyPs in the regulatory processes in bacterial cells: (→) biosyn-
thesis of compounds; (--->) utilization of compounds; (— ) regulatory effects.
Yeast showed an accumulation of PolyP 3 following PolyP hydrolysis induced by amines
and basic amino acids (Greenfeld et al., 1987). The degradation of ‘NMR-observable’,
probably vacuolar, PolyP to short-chain polymers in the cells of Chemostat-cultivated
S. cerevisiae contributed to neutralizing the added alkalinity (Castro et al., 1995, 1999). In
contrast, when the vacuolar vph1-1 mutant, lacking ‘NMR-visible’ PolyP, was subjected
to alkalinization, the absence of a vacuolar source of phosphate slowed re-acidification
(Castro et al., 1999). Anaerobiosis resulted in the complete hydrolysis of PolyP to P i
(Castro et al., 1995).