Page 230 - Applied Probability
P. 230
10. Molecular Phylogeny
216
tidy classification by comparing 16s ribosomal RNA sequences from a va-
riety of representative eukaryotic and prokaryotic organisms. His analysis
refutes the archebacterial grouping and supports the eocytes as the closest
bacterial ancestor of the eukaryotes.
In this example we examine a small portion of Lake’s original data. The
relevant subset consists of 1,092 aligned bases from the rRNA of the or-
ganisms A. salina (a eukaryote), B. subtilis (a eubacterium), H. morrhuae
(a halobacterium), and D. mobilis (an eocyte). These four taxa can be
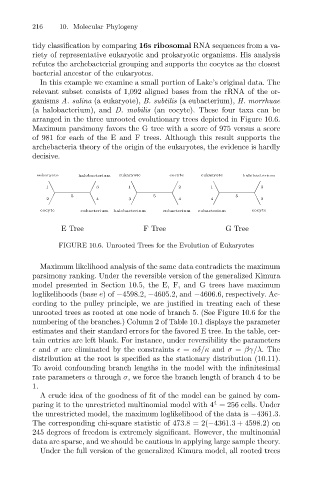
arranged in the three unrooted evolutionary trees depicted in Figure 10.6.
Maximum parsimony favors the G tree with a score of 975 versus a score
of 981 for each of the E and F trees. Although this result supports the
archebacteria theory of the origin of the eukaryotes, the evidence is hardly
decisive.
eukaryote halobacterium eukaryote eocyte eukaryote halobacterium
1 3 1 2 1 3
2 5 4 3 5 4 4 5 2
eocyte eubacterium halobacterium eubacterium eubacterium eocyte
E Tree F Tree G Tree
FIGURE 10.6. Unrooted Trees for the Evolution of Eukaryotes
Maximum likelihood analysis of the same data contradicts the maximum
parsimony ranking. Under the reversible version of the generalized Kimura
model presented in Section 10.5, the E, F, and G trees have maximum
loglikelihoods (base e)of −4598.2, −4605.2, and −4606.6, respectively. Ac-
cording to the pulley principle, we are justified in treating each of these
unrooted trees as rooted at one node of branch 5. (See Figure 10.6 for the
numbering of the branches.) Column 2 of Table 10.1 displays the parameter
estimates and their standard errors for the favored E tree. In the table, cer-
tain entries are left blank. For instance, under reversibility the parameters
and σ are eliminated by the constraints = αδ/κ and σ = βγ/λ. The
distribution at the root is specified as the stationary distribution (10.11).
To avoid confounding branch lengths in the model with the infinitesimal
rate parameters α through σ, we force the branch length of branch 4 to be
1.
A crude idea of the goodness of fit of the model can be gained by com-
4
paring it to the unrestricted multinomial model with 4 = 256 cells. Under
the unrestricted model, the maximum loglikelihood of the data is −4361.3.
The corresponding chi-square statistic of 473.8= 2(−4361.3 + 4598.2) on
245 degrees of freedom is extremely significant. However, the multinomial
data are sparse, and we should be cautious in applying large sample theory.
Under the full version of the generalized Kimura model, all rooted trees