Page 235 - Applied Probability
P. 235
10. Molecular Phylogeny
is
e
θ 0 c i
min{r,m−i} θ j 1 {c i =c i+j }
j=1
so forth until a single array holding L c i
formed. Multiplication of this array against a i−1 (c i ,...,c min{i−1+r,m} ) can
be carried out simultaneously with addition over the index c i . When the
is brought into play, its obvious symmetries can be ex-
array e
θ j 1 {c i =c i+j }
ploited to reduce the overall computational burden. The computational
complexity of the forward algorithm scales linearly in m. e 221
These technical details fail to specify how we distinguish between slow
and fast-evolving codon sites. One obvious choice is to again modulate
the rate of evolution through the acceptance probabilities. Slow evolution
can be distinguished from fast evolution by introducing a multiplicative
parameter η ∈ (0, 1) and replacing each acceptance probability ρ i by ηρ i .
For synonymous codon changes, it makes sense to retain the acceptance
probability of 1.
root
9
7
8
10
6
5
1 2 3 4
rabbit rat goat lemur man opossum
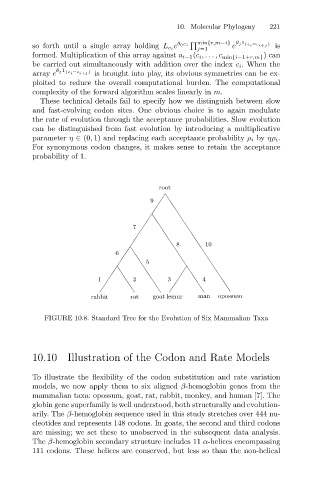
FIGURE 10.8. Standard Tree for the Evolution of Six Mammalian Taxa
10.10 Illustration of the Codon and Rate Models
To illustrate the flexibility of the codon substitution and rate variation
models, we now apply them to six aligned β-hemoglobin genes from the
mammalian taxa: opossum, goat, rat, rabbit, monkey, and human [7]. The
globin gene superfamily is well understood, both structurally and evolution-
arily. The β-hemoglobin sequence used in this study stretches over 444 nu-
cleotides and represents 148 codons. In goats, the second and third codons
are missing; we set these to unobserved in the subsequent data analysis.
The β-hemoglobin secondary structure includes 11 α-helices encompassing
111 codons. These helices are conserved, but less so than the non-helical