Page 231 - Applied Probability
P. 231
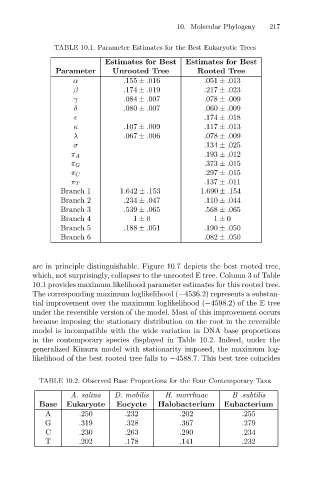
TABLE 10.1. Parameter Estimates for the Best Eukaryotic Trees
Estimates for Best
Rooted Tree
Unrooted Tree
Parameter
.155 ± .016
α
.051 ± .013
β Estimates for Best 10. Molecular Phylogeny 217
.217 ± .023
.174 ± .019
γ .084 ± .007 .078 ± .009
δ .080 ± .007 .060 ± .009
.174 ± .018
κ .107 ± .009 .117 ± .013
λ .067 ± .006 .078 ± .009
σ .134 ± .025
π A .193 ± .012
π G .373 ± .015
.297 ± .015
π C
.137 ± .011
π T
Branch 1 1.642 ± .153 1.690 ± .154
Branch 2 .234 ± .047 .110 ± .044
Branch 3 .539 ± .065 .568 ± .065
Branch 4 1 ± 0 1 ± 0
Branch 5 .188 ± .051 .190 ± .050
Branch 6 .082 ± .050
are in principle distinguishable. Figure 10.7 depicts the best rooted tree,
which, not surprisingly, collapses to the unrooted E tree. Column 3 of Table
10.1 provides maximum likelihood parameter estimates for this rooted tree.
The corresponding maximum loglikelihood (−4536.2) represents a substan-
tial improvement over the maximum loglikelihood (−4598.2) of the E tree
under the reversible version of the model. Most of this improvement occurs
because imposing the stationary distribution on the root in the reversible
model is incompatible with the wide variation in DNA base proportions
in the contemporary species displayed in Table 10.2. Indeed, under the
generalized Kimura model with stationarity imposed, the maximum log-
likelihood of the best rooted tree falls to −4588.7. This best tree coincides
TABLE 10.2. Observed Base Proportions for the Four Contemporary Taxa
A. salina D. mobilis H. morrhuae B .subtilis
Base Eukaryote Eocycte Halobacterium Eubacterium
A .250 .232 .202 .255
G .319 .328 .367 .279
C .230 .263 .290 .234
T .202 .178 .141 .232