Page 48 - Applied Probability
P. 48
2. Counting Methods and the EM Algorithm
likelihood estimate ˆ p = .2679 is consistent with the theoretical value of
p =1/4 for an autosomal recessive. Starting from p = π =1/2, the EM
algorithm takes about 20 iterations to converge to the maximum likelihood
estimates ˆ p = .2679 and ˆ π = .3594.
2.7 Binding Domain Identification 31
Lawrence and Reilly [8] discuss an EM algorithm for recognizing DNA pro-
tein binding domains. Protein binding is intimately connected with regula-
tion of DNA transcription as discussed in the first two sections of Appendix
A. For instance, promoter domains facilitate the binding of transcription
factors that collectively form a complex initiating RNA synthesis by RNA
polymerase II. The DNA bases occuring within a domain are not absolutely
fixed. The famous TATA box about 10 bases upstream of many genes has
the consensus sequence TAxxxT, where the x bases are variable. This vari-
ability makes domain recognition difficult.
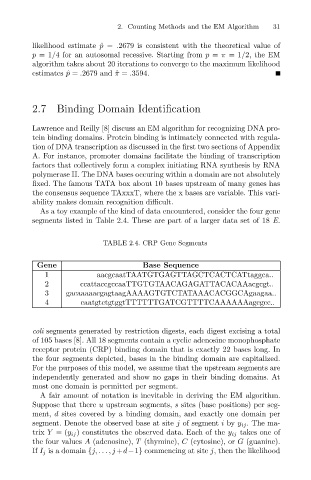
As a toy example of the kind of data encountered, consider the four gene
segments listed in Table 2.4. These are part of a larger data set of 18 E.
TABLE 2.4. CRP Gene Segments
Gene Base Sequence
1 aacgcaatTAATGTGAGTTAGCTCACTCATtaggca..
2 ccattaccgccaaTTGTGTAACAGAGATTACACAAacgcgt..
3 gacaaaaacgagtaagAAAAGTGTCTATAAACACGGCAgaagaa..
4 caatgtctgtggtTTTTTTGATCGTTTTCAAAAAAagcgcc..
coli segments generated by restriction digests, each digest excising a total
of 105 bases [8]. All 18 segments contain a cyclic adenosine monophosphate
receptor protein (CRP) binding domain that is exactly 22 bases long. In
the four segments depicted, bases in the binding domain are capitalized.
For the purposes of this model, we assume that the upstream segments are
independently generated and show no gaps in their binding domains. At
most one domain is permitted per segment.
A fair amount of notation is inevitable in deriving the EM algorithm.
Suppose that there u upstream segments, s sites (base positions) per seg-
ment, d sites covered by a binding domain, and exactly one domain per
segment. Denote the observed base at site j of segment i by y ij . The ma-
trix Y =(y ij ) constitutes the observed data. Each of the y ij takes one of
the four values A (adenosine), T (thymine), C (cytosine), or G (guanine).
If I j is a domain {j, ...,j +d−1} commencing at site j, then the likelihood