Page 140 - Artificial Intelligence for Computational Modeling of the Heart
P. 140
112 Chapter 3 Learning cardiac anatomy
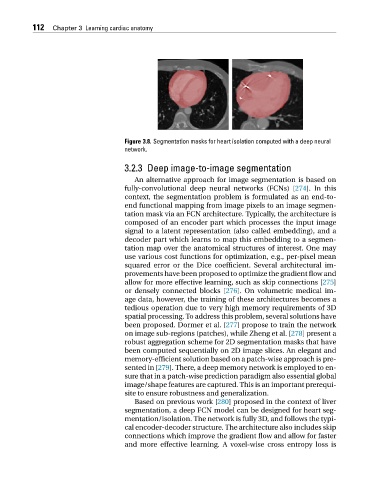
Figure 3.8. Segmentation masks for heart isolation computed with a deep neural
network.
3.2.3 Deep image-to-image segmentation
An alternative approach for image segmentation is based on
fully-convolutional deep neural networks (FCNs) [274]. In this
context, the segmentation problem is formulated as an end-to-
end functional mapping from image pixels to an image segmen-
tation mask via an FCN architecture. Typically, the architecture is
composed of an encoder part which processes the input image
signal to a latent representation (also called embedding), and a
decoder part which learns to map this embedding to a segmen-
tation map over the anatomical structures of interest. One may
use various cost functions for optimization, e.g., per-pixel mean
squared error or the Dice coefficient. Several architectural im-
provements have been proposed to optimize the gradient flow and
allow for more effective learning, such as skip connections [275]
or densely connected blocks [276]. On volumetric medical im-
age data, however, the training of these architectures becomes a
tedious operation due to very high memory requirements of 3D
spatial processing. To address this problem, several solutions have
been proposed. Dormer et al. [277] propose to train the network
on image sub-regions (patches), while Zheng et al. [278]present a
robust aggregation scheme for 2D segmentation masks that have
been computed sequentially on 2D image slices. An elegant and
memory-efficient solution based on a patch-wise approach is pre-
sented in [279]. There, a deep memory network is employed to en-
sure that in a patch-wise prediction paradigm also essential global
image/shape features are captured. This is an important prerequi-
site to ensure robustness and generalization.
Based on previous work [280] proposed in the context of liver
segmentation, a deep FCN model can be designed for heart seg-
mentation/isolation. The network is fully 3D, and follows the typi-
cal encoder-decoder structure. The architecture also includes skip
connections which improve the gradient flow and allow for faster
and more effective learning. A voxel-wise cross entropy loss is