Page 135 - Artificial Intelligence for Computational Modeling of the Heart
P. 135
Chapter 3 Learning cardiac anatomy 107
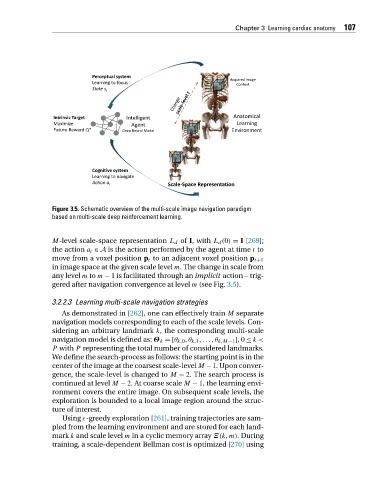
Figure 3.5. Schematic overview of the multi-scale image navigation paradigm
based on multi-scale deep reinforcement learning.
M-level scale-space representation L d of I, with L d (0) = I [269];
the action a t ∈ A is the action performed by the agent at time t to
move from a voxel position p t to an adjacent voxel position p t+1
in image space at the given scale level m. The change in scale from
any level m to m − 1 is facilitated through an implicit action – trig-
gered after navigation convergence at level m (see Fig. 3.5).
3.2.2.3 Learning multi-scale navigation strategies
As demonstrated in [262], one can effectively train M separate
navigation models corresponding to each of the scale levels. Con-
sidering an arbitrary landmark k, the corresponding multi-scale
navigation model is defined as: Θ k =[θ k,0 ,θ k,1 ,...,θ k,M−1 ], 0 ≤ k<
P with P representing the total number of considered landmarks.
We define the search-process as follows: the starting point is in the
center of the image at the coarsest scale-level M −1.Uponconver-
gence, the scale-level is changed to M − 2. The search process is
continued at level M − 2. At coarse scale M − 1, the learning envi-
ronment covers the entire image. On subsequent scale levels, the
exploration is bounded to a local image region around the struc-
ture of interest.
Using -greedy exploration [261], training trajectories are sam-
pled from the learning environment and are stored for each land-
mark k and scale level m in a cyclic memory array Ξ(k,m).During
training, a scale-dependent Bellman cost is optimized [270]using