Page 130 - Artificial Intelligence for Computational Modeling of the Heart
P. 130
102 Chapter 3 Learning cardiac anatomy
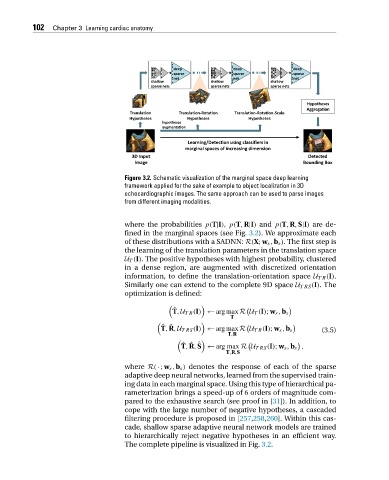
Figure 3.2. Schematic visualization of the marginal space deep learning
framework applied for the sake of example to object localization in 3D
echocardiographic images. The same approach can be used to parse images
from different imaging modalities.
where the probabilities p(T|I), p(T,R|I) and p(T,R,S|I) are de-
fined in the marginal spaces (see Fig. 3.2). We approximate each
of these distributions with a SADNN: R(X;w s ,b s ). The first step is
the learning of the translation parameters in the translation space
U T (I). The positive hypotheses with highest probability, clustered
in a dense region, are augmented with discretized orientation
information, to define the translation-orientation space U TR (I).
Similarly one can extend to the complete 9D space U TRS (I).The
optimization is defined:
ˆ
T,U TR (I) ← argmaxR U T (I);w s ,b s
T
ˆ ˆ
T,R,U TRS (I) ← argmaxR U TR (I);w s ,b s (3.5)
T,R
ˆ ˆ ˆ
T,R,S ← arg max R U TRS (I);w s ,b s ,
T,R,S
where R(·;w s ,b s ) denotes the response of each of the sparse
adaptive deep neural networks, learned from the supervised train-
ing data in each marginal space. Using this type of hierarchical pa-
rameterization brings a speed-up of 6 orders of magnitude com-
pared to the exhaustive search (see proof in [31]). In addition, to
cope with the large number of negative hypotheses, a cascaded
filtering procedure is proposed in [257,258,260]. Within this cas-
cade, shallow sparse adaptive neural network models are trained
to hierarchically reject negative hypotheses in an efficient way.
The complete pipeline is visualized in Fig. 3.2.