Page 56 - Academic Press Encyclopedia of Physical Science and Technology 3rd Molecular Biology
P. 56
P1: GRA Final
Encyclopedia of Physical Science and Technology EN006H-655 June 29, 2001 21:21
Gene Expression, Regulation of 505
to be regulated by inhibitory proteins that interact with
TBP. Such TBP–inhibitory protein complexes may serve
an important regulatory role by keeping genes which have
been removed from inactive chromatin in a repressed but
rapidly inducible state.
1. Regulation of Transcription
by Chromatin Remodeling
During the last decade a wealth of information has demon-
strated the significance of the chromatin template for the
transcriptional activity of a promoter. It has been known
for decades that the DNA in our cells is wrapped around a
protein core called the nucleosome. The nucleosome con-
sists of two copies each of four histones: H2A, H2B, H3,
and H4. The DNA is packaged into either a loose struc-
ture called euchromatin or a more highly ordered structure
called heterochromatin. The heterochromatin fraction is
transcriptionally inactive, whereas active genes are found
in the euchromatin fraction of the DNA.
Histones are modified by acetylation, phosphorylation,
methylation, and ubiqutination. During recent years an
impressive amount of work has demonstrated the sig-
nificance of reversible histone acetylation as a regu-
latory mechanism controlling gene expression. Several
lysines on the amino-terminal tail of each core histone can
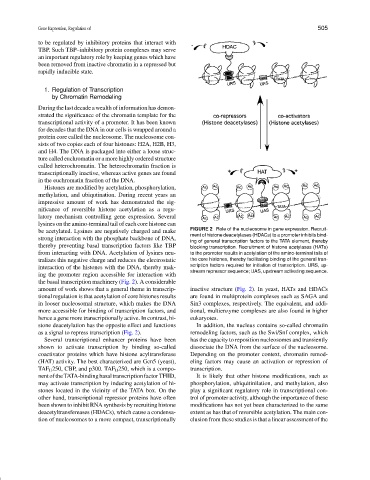
be acetylated. Lysines are negatively charged and make FIGURE 2 Role of the nucleosome in gene expression. Recruit-
ment of histone deacetylases (HDACs) to a promoter inhibits bind-
strong interaction with the phosphate backbone of DNA,
ing of general transcription factors to the TATA element, thereby
thereby preventing basal transcription factors like TBP blocking transcription. Recruitment of histone acetylases (HATs)
from interacting with DNA. Acetylation of lysines neu- to the promoter results in acetylation of the amino-terminal tails of
tralizes this negative charge and reduces the electrostatic the core histones, thereby facilitating binding of the general tran-
interaction of the histones with the DNA, thereby mak- scription factors required for initiation of transcription. URS, up-
stream repressor sequence; UAS, upstream activating sequence.
ing the promoter region accessible for interaction with
the basal transcription machinery (Fig. 2). A considerable
amount of work shows that a general theme in transcrip- inactive structure (Fig. 2). In yeast, HATs and HDACs
tional regulation is that acetylation of core histones results are found in multiprotein complexes such as SAGA and
in looser nucleosomal structure, which makes the DNA Sin3 complexes, respectively. The equivalent, and addi-
more accessible for binding of transcription factors, and tional, multienzyme complexes are also found in higher
hence a gene more transcriptionally active. In contrast, hi- eukaryotes.
stone deacetylation has the opposite effect and functions In addition, the nucleus contains so-called chromatin
as a signal to repress transcription (Fig. 2). remodeling factors, such as the Swi/Snf complex, which
Several transcriptional enhancer proteins have been has the capacity to reposition nucleosomes and transiently
shown to activate transcription by binding so-called dissociate the DNA from the surface of the nucleosome.
coactivator proteins which have histone acyltransferase Depending on the promoter context, chromatin remod-
(HAT) activity. The best characterized are Gcn5 (yeast), eling factors may cause an activation or repression of
TAF II 250, CBP, and p300. TAF II 250, which is a compo- transcription.
nentoftheTATA-bindingbasaltranscriptionfactorTFIID, It is likely that other histone modifications, such as
may activate transcription by inducing acetylation of hi- phosphorylation, ubiquitinilation, and methylation, also
stones located in the vicinity of the TATA box. On the play a significant regulatory role in transcriptional con-
other hand, transcriptional repressor proteins have often trol of promoter activity, although the importance of these
been shown to inhibit RNA synthesis by recruiting histone modifications has not yet been characterized to the same
deacetyltransfereases (HDACs), which cause a condensa- extent as has that of reversible acetylation. The main con-
tion of nucleosomes to a more compact, transcriptionally clusion from these studies is that a linear assessment of the