Page 59 - Academic Press Encyclopedia of Physical Science and Technology 3rd Molecular Biology
P. 59
P1: GRA Final
Encyclopedia of Physical Science and Technology EN006H-655 June 29, 2001 21:21
508 Gene Expression, Regulation of
and tRNA. The core polymerase is a four-subunit enzyme
consisting of two a, one b, and one b subunit. However,
the holoenzyme, which is the complete enzyme, contains
the core polymerase plus the sigma factor, which may
regarded as the prokaryotic equivalent of the general tran-
scription factors found in eukaryotes. The sigma factor is
required for proper RNA polymerase binding to a prokary-
otic promoter. After initiation of transcription the sigma
factor leaves the polymerase complex and elongation is
taken care of by the core polymerase.
Similartoaeukaryoticpromoteraprototypicalprokary-
otic core promoter contains a conserved TATAAT located
at position −10 relative to the transcription start site and
resembling the eukaryotic TATA element. In addition,
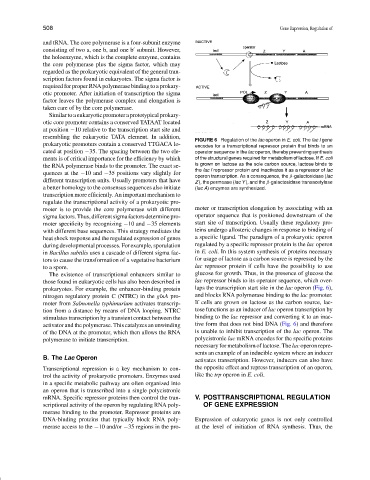
FIGURE 6 Regulation of the lac operon in E. coli. The lac I gene
prokaryotic promoters contain a conserved TTGACA lo- encodes for a transcriptional repressor protein that binds to an
cated at position −35. The spacing between the two ele- operator sequence in the lac operon, thereby preventing synthesis
ments is of critical importance for the efficiency by which of the structural genes required for metabolism of lactose. If E. coli
the RNA polymerase binds to the promoter. The exact se- is grown on lactose as the sole carbon source, lactose binds to
the lac I repressor protein and inactivates it as a repressor of lac
quences at the −10 and −35 positions vary slightly for
operon transcription. As a consequence, the β-galactosidase (lac
different transcription units. Usually promoters that have Z ), the permease (lac Y ), and the β-galactosidase transacetylase
a better homology to the consensus sequences also initiate (lac A) enzymes are synthesized.
transcription more efficiently. An important mechanism to
regulate the transcriptional activity of a prokaryotic pro-
moter is to provide the core polymerase with different moter or transcription elongation by associating with an
sigmafactors.Thus,differentsigmafactorsdeterminepro- operator sequence that is positioned downstream of the
moter specificity by recognizing −10 and −35 elements start site of transcription. Usually these regulatory pro-
with different base sequences. This strategy mediates the teins undergo allosteric changes in response to binding of
heat shock response and the regulated expression of genes a specific ligand. The paradigm of a prokaryotic operon
during developmental processes. For example, sporulation regulated by a specific repressor protein is the lac operon
in Bacillus subtilis uses a cascade of different sigma fac- in E. coli. In this system synthesis of proteins necessary
tors to cause the transformation of a vegetative bacterium for usage of lactose as a carbon source is repressed by the
to a spore. lac repressor protein if cells have the possibility to use
The existence of transcriptional enhancers similar to glucose for growth. Thus, in the presence of glucose the
those found in eukaryotic cells has also been described in lac repressor binds to its operator sequence, which over-
prokaryotes. For example, the enhancer-binding protein laps the transcription start site in the lac operon (Fig. 6),
nitrogen regulatory protein C (NTRC) in the glnA pro- and blocks RNA polymerase binding to the lac promoter.
moter from Salmonella typhimurium activates transcrip- If cells are grown on lactose as the carbon source, lac-
tion from a distance by means of DNA looping. NTRC tose functions as an inducer of lac operon transcription by
stimulates transcription by a transient contact between the binding to the lac repressor and converting it to an inac-
activator and the polymerase. This catalyzes an unwinding tive form that does not bind DNA (Fig. 6) and therefore
of the DNA at the promoter, which then allows the RNA is unable to inhibit transcription of the lac operon. The
polymerase to initiate transcription. polycistronic lac mRNA encodes for the specific proteins
necessary for metabolism of lactose. The lac operon repre-
sents an example of an inducible system where an inducer
B. The Lac Operon activates transcription. However, inducers can also have
Transcriptional repression is a key mechanism to con- the opposite effect and repress transcription of an operon,
trol the activity of prokaryotic promoters. Enzymes used like the trp operon in E. coli.
in a specific metabolic pathway are often organized into
an operon that is transcribed into a single polycistronic
mRNA. Specific repressor proteins then control the tran- V. POSTTRANSCRIPTIONAL REGULATION
scriptional activity of the operon by regulating RNA poly- OF GENE EXPRESSION
merase binding to the promoter. Repressor proteins are
DNA-binding proteins that typically block RNA poly- Expression of eukaryotic genes is not only controlled
merase access to the −10 and/or −35 regions in the pro- at the level of initiation of RNA synthesis. Thus, the