Page 54 - Academic Press Encyclopedia of Physical Science and Technology 3rd Molecular Biology
P. 54
P1: GRA Final
Encyclopedia of Physical Science and Technology EN006H-655 June 29, 2001 21:21
Gene Expression, Regulation of 503
elongation, during which the RNA polymerase, by a pro-
cessive mechanism, moves along the DNA template in
a5 -to-3 direction and extends the growing RNA chain
by copying one nucleotide at a time; and (3) termination,
where RNA synthesis ends and the RNA polymerase com-
plex disassembles from the transcription unit.
Because of space limitation this review will mostly
cover RNA synthesis and maturation of protein-encoding
messenger RNAs (mRNAs). However, similar mecha-
nisms are used to regulate synthesis of other types of RNA
molecules.
III. REGULATION OF TRANSCRIPTION
IN EUKARYOTES
EukaryoticcellscontainthreeDNA-dependentRNApoly-
merases which are responsible for synthesis of specific
RNA molecules in the cell. RNA polymerase I is respon-
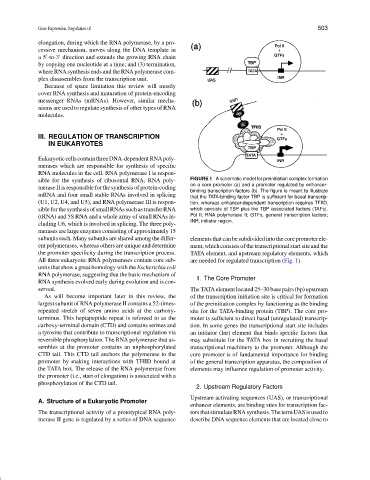
sible for the synthesis of ribosomal RNA, RNA poly- FIGURE 1 A schematic model for preinitiation complex formation
on a core promoter (a) and a promoter regulated by enhancer-
merase II is responsible for the synthesis of protein-coding
binding transcription factors (b). The figure is meant to illustrate
mRNA and four small stable RNAs involved in splicing that the TATA-binding factor TBP is sufficient for basal transcrip-
(U1, U2, U4, and U5), and RNA polymerase III is respon- tion, whereas enhancer-dependent transcription requires TFIID,
sible for the synthesis of small RNAs such as transfer RNA which consists of TBP plus the TBP associated factors (TAFs).
(tRNA) and 5S RNA and a whole array of small RNAs in- Pol II, RNA polymerase II; GTFs, general transcription factors;
INR, initiator region.
cluding U6, which is involved in splicing. The three poly-
merases are large enzymes consisting of approximately 15
subunits each. Many subunits are shared among the differ- elements that can be subdivided into the core promoter ele-
ent polymerases, whereas others are unique and determine ment,whichconsistsofthetranscriptionalstartsiteandthe
the promoter specificity during the transcription process. TATA element, and upstream regulatory elements, which
All three eukaryotic RNA polymerases contain core sub- are needed for regulated transcription (Fig. 1).
units that show a great homology with the Escherichia coli
RNA polymerase, suggesting that the basic mechanism of
1. The Core Promoter
RNA synthesis evolved early during evolution and is con-
served. The TATA element located 25–30 base pairs (bp) upstream
As will become important later in this review, the of the transcription initiation site is critical for formation
largest subunit of RNA polymerase II contains a 52-times- of the preinitiation complex by functioning as the binding
repeated stretch of seven amino acids at the carboxy- site for the TATA-binding protein (TBP). The core pro-
terminus. This heptapeptide repeat is refereed to as the moter is sufficient to direct basal (unregulated) transcrip-
carboxy-terminal domain (CTD) and contains serines and tion. In some genes the transcriptional start site includes
a tyrosine that contribute to transcriptional regulation via an initiator (Inr) element that binds specific factors that
reversible phosphorylation. The RNA polymerase that as- may substitute for the TATA box in recruiting the basal
sembles at the promoter contains an unphosphorylated transcriptional machinery to the promoter. Although the
CTD tail. This CTD tail anchors the polymerase to the core promoter is of fundamental importance for binding
promoter by making interactions with TFIID bound at of the general transcription apparatus, the composition of
the TATA box. The release of the RNA polymerase from elements may influence regulation of promoter activity.
the promoter (i.e., start of elongation) is associated with a
phosphorylation of the CTD tail.
2. Upstream Regulatory Factors
Upstream activating sequences (UAS), or transcriptional
A. Structure of a Eukaryotic Promoter
enhancer elements, are binding sites for transcription fac-
The transcriptional activity of a prototypical RNA poly- tors that stimulate RNA synthesis. The term UAS is used to
merase II gene is regulated by a series of DNA sequence describe DNA sequence elements that are located close to