Page 50 - Handbook of Deep Learning in Biomedical Engineering Techniques and Applications
P. 50
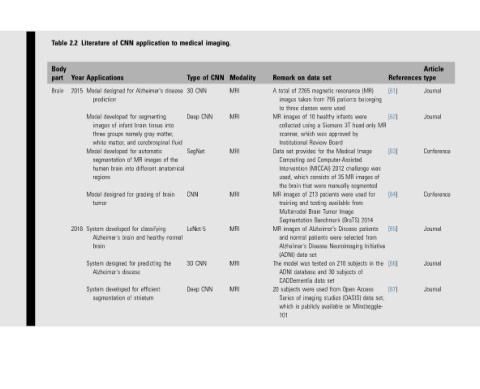
Article type Journal Journal Conference Conference Journal Journal Journal
References
[61]
[62]
MR [63] was of [64] [65] Initiative [66] the [67] set,
(MR) belonging were head-only Image images segmented for used 2014 patients from in of Access data Mindboggle-
resonance patients used infants 3T by approved Medical Computer-Assisted challenge 2012 MR 35 manually were from available Image (BraTS) Disease selected Neuroimaging subjects 210 subjects Open (OASIS) on
set magnetic 755 were healthy Siemens was Board the for (MICCAI) of were patients Tumor Benchmark Alzheimer's were on 30 and set from used studies available
data from classes 10 a using which Review and consists that 213 testing Brain patients Disease set tested data were imaging publicly
on 2265 of taken three of Institutional provided Intervention which brain of and Multimodal Segmentation of normal Alzheimer's data was database CADDementia of is
Remark total A images to images MR collected scanner, set Data Computing used, the images MR training images MR and (ADNI) model The ADNI subjects 20 Series which 101
Modality MRI MRI MRI MRI MRI MRI MRI
imaging. CNN of CNN CNN
medical Type CNN 3D Deep SegNet CNN LeNet-5 CNN 3D Deep
to disease into matter, fluid the of anatomical brain normal the
application Alzheimer's segmenting tissue brain gray cerebrospinal automatic images different of grading classifying healthy and predicting efficient striatum
CNN for for infant namely and for MR of into for for brain for disease for of
of designed developed of groups matter, developed segmentation brain designed developed Alzheimer's designed Alzheimer's developed segmentation
Literature Applications Model prediction Model images three white Model human regions Model tumor System brain System System
2.2 Year 2015 2016
Table Body part Brain