Page 51 - Handbook of Deep Learning in Biomedical Engineering Techniques and Applications
P. 51
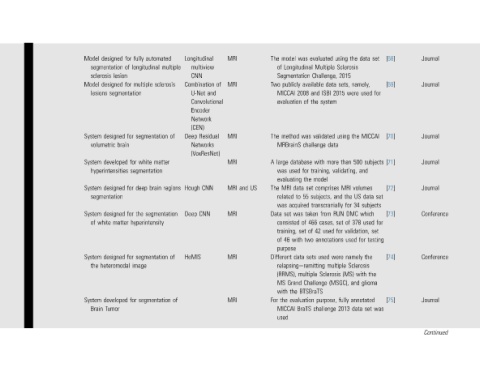
Journal Journal Journal Journal Journal Conference Conference Journal Continued
[68] [69] [70] [71] [72] [73] [74] [75]
set for MICCAI subjects set subjects for set testing the was
data namely, used and volumes data which used for the Sclerosis with glioma annotated set
the Sclerosis were the 500 US the 34 for DMC 378 of validation, used namely (MS) and data
using 2015 sets, data 2015 using data than more validating, MRI and RUN set for were multiple (MSGC), fully 2013
evaluated Multiple Challenge, available ISBI and system the validated challenge with training, model comprises subjects, transcranially from cases, 466 used 42 annotations used Sclerosis Challenge purpose, challenge
was Longitudinal 2008 of was database for the set data 55 to acquired taken was of of set two with sets data relapsingeremitting multiple BTSBraTS evaluation BraTS
model Segmentation publicly MICCAI evaluation method MRBrainS large used was evaluating MRI related was set consisted training, 46 purpose (RRMS), Grand the with the MICCAI used
The of Two The A The Data of Different MS For
US
and
MRI MRI MRI MRI MRI MRI MRI MRI
of and Residual
Longitudinal multiview CNN Combination U-Net Convolutional Encoder Network (CEN) Deep Networks (VoxResNet) CNN Hough CNN Deep HeMIS
multiple sclerosis of regions of of
automated segmentation matter brain segmentation segmentation segmentation
fully longitudinal multiple white for segmentation deep the hyperintensity image for
for of for segmentation for brain for for matter for
designed segmentation lesion designed designed volumetric developed hyperintensities designed segmentation designed white designed heteromodal developed Tumor
Model sclerosis Model lesions System System System System of System the System Brain