Page 118 - Macromolecular Crystallography
P. 118
MOLECULAR REPLACEMENT TECHNIQUES 107
PSI-BLAST
CLUSTAL-W
T-Coffee
Diffraction data
Model(s) cell parameters Sequences
# mol. per a.u.
Modeller Truncated
Whatif model(s)
elNemo
Molecular
replacement:
– AMoRe
– EPMR...
Refinement:
– Rigid body
– Normal modes
– Sim. annealing
Automatic
construction:
– ARP/wARP
– Solve/resolve
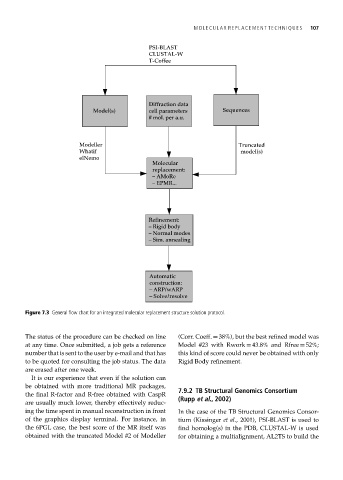
Figure 7.3 General flow chart for an integrated molecular replacement structure solution protocol.
The status of the procedure can be checked on line (Corr. Coeff. = 38%), but the best refined model was
at any time. Once submitted, a job gets a reference Model #23 with Rwork = 43.8% and Rfree = 52%;
number that is sent to the user by e-mail and that has this kind of score could never be obtained with only
to be quoted for consulting the job status. The data Rigid Body refinement.
are erased after one week.
It is our experience that even if the solution can
be obtained with more traditional MR packages,
7.9.2 TB Structural Genomics Consortium
the final R-factor and R-free obtained with CaspR
(Rupp et al., 2002)
are usually much lower, thereby effectively reduc-
ing the time spent in manual reconstruction in front In the case of the TB Structural Genomics Consor-
of the graphics display terminal. For instance, in tium (Kissinger et al., 2001), PSI-BLAST is used to
the 6PGL case, the best score of the MR itself was find homolog(s) in the PDB, CLUSTAL-W is used
obtained with the truncated Model #2 of Modeller for obtaining a multialignment, AL2TS to build the