Page 235 - Advanced Organic Chemistry Part A - Structure and Mechanisms, 5th ed (2007) - Carey _ Sundberg
P. 235
The strength of CE as an analytical tool is the very high degree of enantioselection 215
that can be achieved, along with high speed and sensitivity. It is more difficult to use
CE on a preparative scale, although successful separation has been reported on the TOPIC 2.2
milligram scale. 204 Enzymatic Resolution
and Desymmetrization
We have seen that in each of these means of enantiomeric separation, chiral
recognition depends upon a combination of intermolecular forces, including electro-
static attractions, hydrogen bonding, and - stacking. These differential interactions
then lead to distinctions between the properties of the two enantiomers, such as
chemical shifts in NMR methods or relative mobility in chiral chromatography and
electrophoresis. There is much current interest in both the analysis of these interactions
and manipulation of structure to increase selectivity.
Topic 2.2. Enzymatic Resolution and Desymmetrization
Enzymatic resolution is based on the ability of enzymes (catalytic proteins) to
distinguish between R- and S-enantiomers or between enantiotopic pro-R and pro-
S groups in prochiral compounds. 205 The selective conversion of pro-R and pro-S
groups is often called desymmetrization or asymmetrization. 206 Note that in contrast to
enzymatic resolution, which can at best provide half the racemic product as resolved
material, prochiral compounds can be completely converted to a single enantiomer,
provided that the selectivity is high enough. Complete conversion of a racemic mixture
to a single enantiomeric product can sometimes be accomplished by coupling an
enzymatic resolution with another reaction (chemical or enzymatic) that racemizes the
reactant. This is called dynamic resolution, 207 and it has been accomplished for several
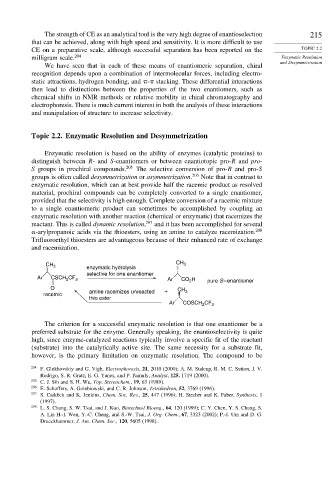
-arylpropanoic acids via the thioesters, using an amine to catalyze racemization. 208
Trifluoroethyl thioesters are advantageous because of their enhanced rate of exchange
and racemization.
CH 3 enzymatic hydrolysis CH 3
selective for one enantiomer
Ar CSCH CF 3 Ar CO H pure S – enantiomer
2
2
O + CH
racemic amine racemizes unreacted 3
thio ester
Ar COSCH CF 3
2
The criterion for a successful enzymatic resolution is that one enantiomer be a
preferred substrate for the enzyme. Generally speaking, the enantioselectivity is quite
high, since enzyme-catalyzed reactions typically involve a specific fit of the reactant
(substrate) into the catalytically active site. The same necessity for a substrate fit,
however, is the primary limitation on enzymatic resolution. The compound to be
204 F. Glukhovskiy and G. Vigh, Electrophoresis, 21, 2010 (2000); A. M. Stalcup, R. M. C. Sutton, J. V.
Rodrigo, S. R. Gratz, E. G. Yanes, and P. Painuly, Analyst, 125, 1719 (2000).
205 C. J. Sih and S. H. Wu, Top. Stereochem., 19, 63 (1989).
206
E. Schoffers, A. Golebiowski, and C. R. Johnson, Tetrahedron, 52, 3769 (1996).
207 S. Caddick and K. Jenkins, Chem. Soc. Rev., 25, 447 (1996); H. Stecher and K. Faber, Synthesis,1
(1997).
208
L. S. Chang, S. W. Tsai, and J. Kuo, Biotechnol Bioeng., 64, 120 (1999); C. Y. Chen, Y. S. Chang, S.
A. Lin H.-I. Wen, Y.-C. Cheng, and S.-W. Tsai, J. Org. Chem., 67, 3323 (2002); P.-J. Um and D. G.
Drueckhammer, J. Am. Chem. Soc., 120, 5605 (1998)..