Page 149 - Artificial Intelligence for Computational Modeling of the Heart
P. 149
Chapter 4 Data-driven reduction of cardiac models 121
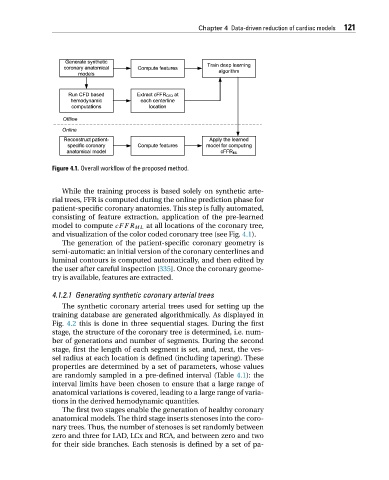
Figure 4.1. Overall workflow of the proposed method.
While the training process is based solely on synthetic arte-
rial trees, FFR is computed during the online prediction phase for
patient-specific coronary anatomies. This step is fully automated,
consisting of feature extraction, application of the pre-learned
model to compute cFFR ML at all locations of the coronary tree,
and visualization of the color coded coronary tree (see Fig. 4.1).
The generation of the patient-specific coronary geometry is
semi-automatic: an initial version of the coronary centerlines and
luminal contours is computed automatically, and then edited by
the user after careful inspection [335]. Once the coronary geome-
try is available, features are extracted.
4.1.2.1 Generating synthetic coronary arterial trees
The synthetic coronary arterial trees used for setting up the
training database are generated algorithmically. As displayed in
Fig. 4.2 this is done in three sequential stages. During the first
stage, the structure of the coronary tree is determined, i.e. num-
ber of generations and number of segments. During the second
stage, first the length of each segment is set, and, next, the ves-
sel radius at each location is defined (including tapering). These
properties are determined by a set of parameters, whose values
are randomly sampled in a pre-defined interval (Table 4.1): the
interval limits have been chosen to ensure that a large range of
anatomical variations is covered, leading to a large range of varia-
tions in the derived hemodynamic quantities.
The first two stages enable the generation of healthy coronary
anatomical models. The third stage inserts stenoses into the coro-
nary trees. Thus, the number of stenoses is set randomly between
zero and three for LAD, LCx and RCA, and between zero and two
for their side branches. Each stenosis is defined by a set of pa-