Page 153 - Artificial Intelligence for Computational Modeling of the Heart
P. 153
Chapter 4 Data-driven reduction of cardiac models 125
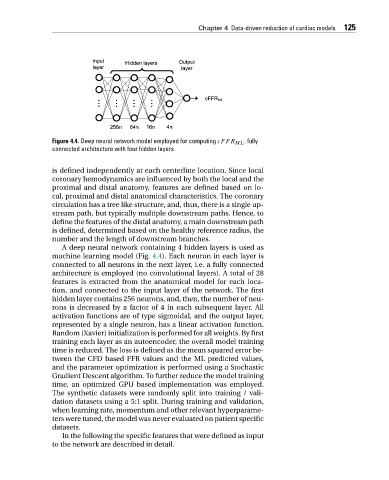
Figure 4.4. Deep neural network model employed for computing cFFR ML : fully
connected architecture with four hidden layers.
is defined independently at each centerline location. Since local
coronary hemodynamics are influenced by both the local and the
proximal and distal anatomy, features are defined based on lo-
cal, proximal and distal anatomical characteristics. The coronary
circulation has a tree like structure, and, thus, there is a single up-
stream path, but typically multiple downstream paths. Hence, to
define the features of the distal anatomy, a main downstream path
is defined, determined based on the healthy reference radius, the
number and the length of downstream branches.
A deep neural network containing 4 hidden layers is used as
machine learning model (Fig. 4.4). Each neuron in each layer is
connected to all neurons in the next layer, i.e. a fully connected
architecture is employed (no convolutional layers). A total of 28
features is extracted from the anatomical model for each loca-
tion, and connected to the input layer of the network. The first
hidden layer contains 256 neurons, and, then, the number of neu-
rons is decreased by a factor of 4 in each subsequent layer. All
activation functions are of type sigmoidal, and the output layer,
represented by a single neuron, has a linear activation function.
Random (Xavier) initialization is performed for all weights. By first
training each layer as an autoencoder, the overall model training
time is reduced. The loss is defined as the mean squared error be-
tween the CFD based FFR values and the ML predicted values,
and the parameter optimization is performed using a Stochastic
Gradient Descent algorithm. To further reduce the model training
time, an optimized GPU based implementation was employed.
The synthetic datasets were randomly split into training / vali-
dation datasets using a 5:1 split. During training and validation,
when learning rate, momentum and other relevant hyperparame-
ters were tuned, the model was never evaluated on patient specific
datasets.
In the following the specific features that were defined as input
to the network are described in detail.