Page 133 - Chiral Separation Techniques
P. 133
110 4 CHIRBASE: Database Current Status and Derived Research Applications using …
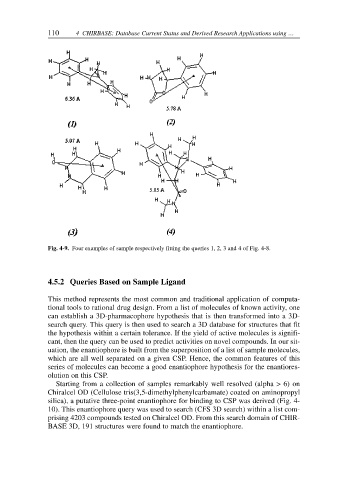
Fig. 4-9. Four examples of sample respectively fitting the queries 1, 2, 3 and 4 of Fig. 4-8.
4.5.2 Queries Based on Sample Ligand
This method represents the most common and traditional application of computa-
tional tools to rational drug design. From a list of molecules of known activity, one
can establish a 3D-pharmacophore hypothesis that is then transformed into a 3D-
search query. This query is then used to search a 3D database for structures that fit
the hypothesis within a certain tolerance. If the yield of active molecules is signifi-
cant, then the query can be used to predict activities on novel compounds. In our sit-
uation, the enantiophore is built from the superposition of a list of sample molecules,
which are all well separated on a given CSP. Hence, the common features of this
series of molecules can become a good enantiophore hypothesis for the enantiores-
olution on this CSP.
Starting from a collection of samples remarkably well resolved (alpha > 6) on
Chiralcel OD (Cellulose tris(3,5-dimethylphenylcarbamate) coated on aminopropyl
silica), a putative three-point enantiophore for binding to CSP was derived (Fig. 4-
10). This enantiophore query was used to search (CFS 3D search) within a list com-
prising 4203 compounds tested on Chiralcel OD. From this search domain of CHIR-
BASE 3D, 191 structures were found to match the enantiophore.