Page 418 -
P. 418
Section 12.3 Registering Deformable Objects 386
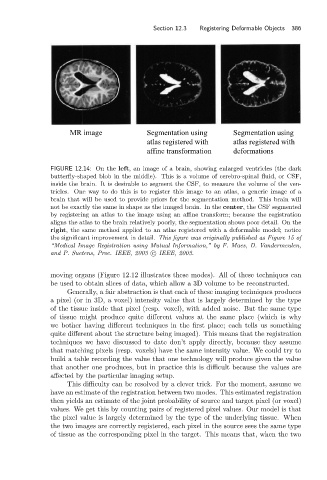
MR image Segmentation using Segmentation using
atlas registered with atlas registered with
affine transformation deformations
FIGURE 12.14: On the left, an image of a brain, showing enlarged ventricles (the dark
butterfly-shaped blob in the middle). This is a volume of cerebro-spinal fluid, or CSF,
inside the brain. It is desirable to segment the CSF, to measure the volume of the ven-
tricles. One way to do this is to register this image to an atlas, a generic image of a
brain that will be used to provide priors for the segmentation method. This brain will
not be exactly the same in shape as the imaged brain. In the center, the CSF segmented
by registering an atlas to the image using an affine transform; because the registration
aligns the atlas to the brain relatively poorly, the segmentation shows poor detail. On the
right, the same method applied to an atlas registered with a deformable model; notice
the significant improvement in detail. This figure was originally published as Figure 15 of
“Medical Image Registration using Mutual Information,” by F. Maes, D. Vandermeulen,
and P. Suetens, Proc. IEEE, 2003 c IEEE, 2003.
moving organs (Figure 12.12 illustrates these modes). All of these techniques can
be used to obtain slices of data, which allow a 3D volume to be reconstructed.
Generally, a fair abstraction is that each of these imaging techniques produces
a pixel (or in 3D, a voxel) intensity value that is largely determined by the type
of the tissue inside that pixel (resp. voxel), with added noise. But the same type
of tissue might produce quite different values at the same place (which is why
we bother having different techniques in the first place; each tells us something
quite different about the structure being imaged). This means that the registration
techniques we have discussed to date don’t apply directly, because they assume
that matching pixels (resp. voxels) have the same intensity value. We could try to
build a table recording the value that one technology will produce given the value
that another one produces, but in practice this is difficult because the values are
affected by the particular imaging setup.
This difficulty can be resolved by a clever trick. For the moment, assume we
have an estimate of the registration between two modes. This estimated registration
then yields an estimate of the joint probability of source and target pixel (or voxel)
values. We get this by counting pairs of registered pixel values. Our model is that
the pixel value is largely determined by the type of the underlying tissue. When
the two images are correctly registered, each pixel in the source sees the same type
of tissue as the corresponding pixel in the target. This means that, when the two