Page 416 -
P. 416
Section 12.3 Registering Deformable Objects 384
the surgeon to the exact position of the tumor inside the patient. Various methods
exist for attaching functional tags to the image of the brain —usually one stimulates
a region of the brain and watches to see what happens — and this information can
also be displayed to the surgeon so that the impact of any damage done can be
minimized. The problem here is pure pose estimation; we need to know the pose
of thebrainimage andthe brainmeasurements withrespect to theperson onthe
table, so that the brain image can be superimposed on the patient in the surgeon’s
display (Figure 12.11).
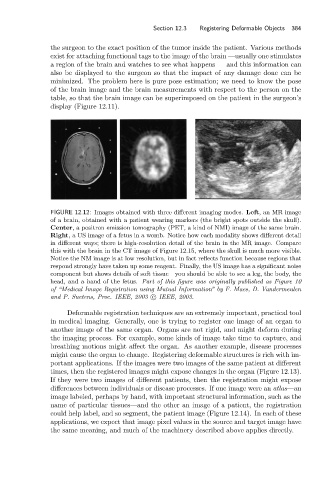
FIGURE 12.12: Images obtained with three different imaging modes. Left, an MR image
of a brain, obtained with a patient wearing markers (the bright spots outside the skull).
Center, a positron emission tomography (PET, a kind of NMI) image of the same brain.
Right, a US image of a fetus in a womb. Notice how each modality shows different detail
in different ways; there is high-resolution detail of the brain in the MR image. Compare
this with the brain in the CT image of Figure 12.15, where the skull is much more visible.
Notice the NM image is at low resolution, but in fact reflects function because regions that
respond strongly have taken up some reagent. Finally, the US image has a significant noise
component but shows details of soft tissue—you should be able to see a leg, the body, the
head, and a hand of the fetus. Part of this figure was originally published as Figure 10
of “Medical Image Registration using Mutual Information” by F. Maes, D. Vandermeulen
and P. Suetens, Proc. IEEE, 2003 c IEEE, 2003.
Deformable registration techniques are an extremely important, practical tool
in medical imaging. Generally, one is trying to register one image of an organ to
another image of the same organ. Organs are not rigid, and might deform during
the imaging process. For example, some kinds of image take time to capture, and
breathing motions might affect the organ. As another example, disease processes
might cause the organ to change. Registering deformable structures is rich with im-
portant applications. If the images were two images of the same patient at different
times, then the registered images might expose changes in the organ (Figure 12.13).
If they were two images of different patients, then the registration might expose
differences between individuals or disease processes. If one image were an atlas—an
image labeled, perhaps by hand, with important structural information, such as the
name of particular tissues—and the other an image of a patient, the registration
could help label, and so segment, the patient image (Figure 12.14). In each of these
applications, we expect that image pixel values in the source and target image have
the same meaning, and much of the machinery described above applies directly.