Page 167 - Academic Press Encyclopedia of Physical Science and Technology 3rd BioTechnology
P. 167
P1: GKY/GLQ/GTK P2: GLM Final Pages
Encyclopedia of Physical Science and Technology en009I-422 July 6, 2001 19:57
Metabolic Engineering 395
ent types of regulation within the cell (see Fig. 5). In recent
years some very powerful techniques have been developed
for quantification of metabolic fluxes and for identifica-
tion of the active metabolic network—often referred to as
metabolic network analysis. Metabolic network analysis
basically consists of two steps:
Identification of the metabolic network structure (or
pathway topology)
Quantification of the fluxes through the branches of the
metabolic network
For identification of the metabolic network structure, one
may gain much information through the extensive bio-
FIGURE 4 Gene shuffling. A family of genes are cleaved by re- chemistry literature and biochemical databases available
striction enzymes, and ligated randomly. The shuffled genes are on the web (see, e.g., www.genome.ad.jp, which gives
cloned into a proper host, and there are selected for desirable
complete metabolic maps with direct links to sequenced
properties. A good selection procedure is clearly essential since
genes and other information about the individual en-
a very high number of shuffled genes may be obtained.
zymes). Thus, there are many reports on the presence
of specific enzyme activities in many different species,
organisms through exploitation of the biodiversity in the and for most industrially important microorganisms the
world will therefore play an important role in the future major metabolic routes have been identified. However, in
of metabolic engineering, especially since the access to a many cases the complete metabolic network structure is
class of genes from different organisms will enable con- not known, i.e., some of the pathways carrying significant
struction of completely new genes through gene shuffling fluxes have not been identified in the microorganism inves-
(see Fig. 4). Thereby it is possible to construct genes tigated. In these cases enzyme assays can be used to con-
that encode proteins with altered properties, and this ap- firm the presence of specific enzymes and determine the
proach may also be applied for obtaining enzymes with cofactor requirements in these pathways, e.g., whether the
improved properties, e.g., improved stability, improved enzyme uses NADH or NADPH as cofactor. Even though
catalytic activity, or improved affinity toward the sub- enzyme assays are valuable for confirming the presence of
strate(s). Another approach is to apply directed evolution active pathways, they are of limited use for identification
of a given gene through error-prone polymerase chain re- of pathways in the studied microorganism. For these pur-
action (PCR), a technique that may also be used in com- poses, isotope-labeled substrates is a powerful tool, and
bination with gene shuffling. Generally these empirical
approaches have shown to be more powerful than pro-
tein engineering. In the future, when our understanding
between the primary structure of proteins and their func-
tion has been improved, directed modifications of proteins
through protein engineering may, however, enable con-
struction of tailor-made enzymes that may contain desir-
able properties. Thus, enzymes that have increased affin-
ity for a branch point metabolite may be constructed, and
hereby more carbon may be directed toward the end prod-
uct of interest (see discussion later).
IV. METABOLIC NETWORK ANALYSIS
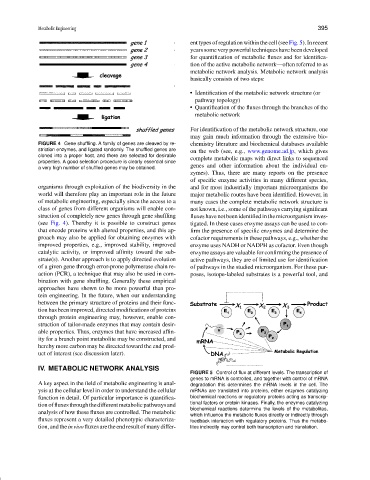
FIGURE 5 Control of flux at different levels. The transcription of
genes to mRNA is controlled, and together with control of mRNA
A key aspect in the field of metabolic engineering is anal- degradation this determines the mRNA levels in the cell. The
ysis at the cellular level in order to understand the cellular mRNAs are translated into proteins, either enzymes catalyzing
function in detail. Of particular importance is quantifica- biochemical reactions or regulatory proteins acting as transcrip-
tionoffluxesthroughthedifferentmetabolicpathwaysand tional factors or protein kinases. Finally, the enzymes catalyzing
biochemical reactions determine the levels of the metabolites,
analysis of how these fluxes are controlled. The metabolic
which influence the metabolic fluxes directly or indirectly through
fluxes represent a very detailed phenotypic characteriza- feedback interaction with regulatory proteins. Thus the metabo-
tion, and the in vivo fluxes are the end result of many differ- lites indirectly may control both transcription and translation.