Page 172 - Academic Press Encyclopedia of Physical Science and Technology 3rd BioTechnology
P. 172
P1: GKY/GLQ/GTK P2: GLM Final Pages
Encyclopedia of Physical Science and Technology en009I-422 July 6, 2001 19:57
400 Metabolic Engineering
VI. TOOLS FROM FUNCTIONAL GENOMICS become available for gene expression analysis in differ-
ent organisms, and today there are commercial available
The active metabolic network functioning in a given cell chips for E. coli, S. cerevisiae, plant cells, different ani-
is determined by the enzymatic makeup of the cell. Fluxes mal cells (only part of the genome currently covered), and
are therefore indirectly also controlled at the level of tran- human cells (also only with part of the genome currently
scription and translation. In fact, there are several different covered). These chips contain oligonucleotide sequences
levels of control as illustrated in Fig. 5: from sequenced genes and allow rapid identification of
complementary sequences in the sample mRNA pool (see
Fig. 10). DNA arrays produced by Affymetrix are gen-
1. Transcriptional control
erated by a photolitography process, which allows the
2. Control of mRNA degradation
synthesis of large numbers of oligonucleotides directly
3. Translational control
on a solid substrate (see www.affymetrix.com for further
4. Protein activation/inactivation
details). An alternative method for production of DNA ar-
5. Allosteric control of enzymes
rays is spotting of oligonucleotide or cDNA solutions by
a robot to a solid substrate followed by immobilization
Due to this hierarchical control, it is difficult to predict of the oligonucleotide or DNA. This method allows for
the overall consequences of a specific genetic modifica- production of custom-made DNA arrays, and is therefore
tion. Thus, a genetic change may result in altered en- more flexible. Normally it is cDNA that is spotted on these
zyme levels, and thereby the metabolite concentrations custom-designed arrays, and the cDNA may have a length
may change. This may influence regulatory proteins that of several hundred nucleotides. This enables a very good
may lead to a secondary effect, both at the transcriptional hybridization, and it is therefore not necessary to have
level and at the level of the enzymes. Generally, it is dif- more than a single probe for each gene.
ficult to predict all the consequences of a certain spe- Besides DNA arrays for transcription profiling, other
cific genetic change, and it may therefore be necessary tools from functional genomics are valuable in the field
to go through the cycle of metabolic engineering several of metabolic engineering. Using two-dimensional elec-
times. However, with novel analysis techniques developed trophoresis, it is possible to identify the pool of proteins
within the field of functional genomics, a very detailed present in a given cell—often referred to as proteomics—
characterization of the cell can be carried out, and this
may enable a far better design of a metabolic engineering
strategy.
A very powerful analytical technique that has emerged
in recent years is DNA arrays or DNA chips, which en-
ables measurement of the expression profile of all genes
within a genome—often referred to as whole genome
transcriptome analysis. Thus, with one measurement it
is possible to monitor which genes are active at specific
conditions, and especially it is possible to rapidly screen
single deletion mutants. Combination of DNA arrays for
genome wide expression monitoring and bioinformatics
has demonstrated to be very powerful for pathway re-
constitution, and in the future this approach is expected
to speed up the assignment of function to orphan genes.
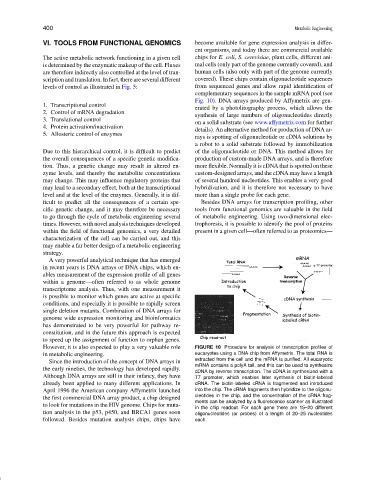
However, it is also expected to play a very valuable role FIGURE 10 Procedure for analysis of transcription profiles of
in metabolic engineering. eucaryotes using a DNA chip from Affymetrix. The total RNA is
extracted from the cell and the mRNA is purified. All eucaryotic
Since the introduction of the concept of DNA arrays in
mRNA contains a polyA tail, and this can be used to synthesize
the early nineties, the technology has developed rapidly.
cDNA by reverse transcription. The cDNA is synthesized with a
Although DNA arrays are still in their infancy, they have T7 promoter, which enables later synthesis of biotin-labeled
already been applied to many different applications. In cRNA. The biotin-labeled cRNA is fragmented and introduced
April 1996 the American company Affymetrix launched into the chip. The cRNA fragments then hybridize to the oligonu-
the first commercial DNA array product, a chip designed cleotides in the chip, and the concentration of the cRNA frag-
ments can be analyzed by a fluorescence scanner as illustrated
to look for mutations in the HIV genome. Chips for muta-
in the chip readout. For each gene there are 15–20 different
tion analysis in the p53, p450, and BRCA1 genes soon oligonucleotides (or probes) of a length of 20–25 nucleotides
followed. Besides mutation analysis chips, chips have each.