Page 166 - Academic Press Encyclopedia of Physical Science and Technology 3rd BioChemistry
P. 166
P1: GPAFinal Pages
Encyclopedia of Physical Science and Technology EN013D-616 July 27, 2001 12:5
206 Protein Structure
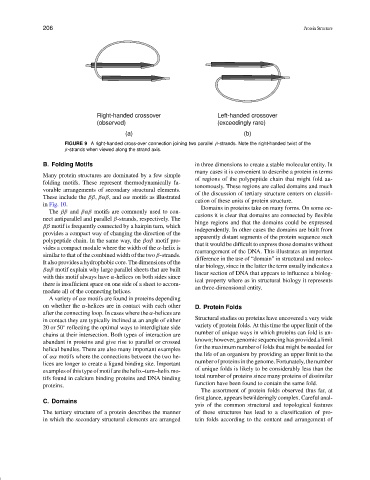
Right-handed crossover Left-handed crossover
(observed) (exceedingly rare)
(a) (b)
FIGURE 9 A right-handed cross-over connection joining two parallel β-strands. Note the right-handed twist of the
β-strands when viewed along the strand axis.
B. Folding Motifs in three dimensions to create a stable molecular entity. In
many cases it is convenient to describe a protein in terms
Many protein structures are dominated by a few simple
of regions of the polypeptide chain that might fold au-
folding motifs. These represent thermodynamically fa-
tonomously. These regions are called domains and much
vorable arrangements of secondary structural elements.
of the discussion of tertiary structure centers on classifi-
These include the ββ, βαβ, and αα motifs as illustrated
cation of these units of protein structure.
in Fig. 10.
Domains in proteins take on many forms. On some oc-
The ββ and βαβ motifs are commonly used to con-
casions it is clear that domains are connected by flexible
nect antiparallel and parallel β-strands, respectively. The
hinge regions and that the domains could be expressed
ββ motif is frequently connected by a hairpin turn, which
independently. In other cases the domains are built from
provides a compact way of changing the direction of the
apparently distant segments of the protein sequence such
polypeptide chain. In the same way, the βαβ motif pro-
that it would be difficult to express those domains without
vides a compact module where the width of the α-helix is
rearrangement of the DNA. This illustrates an important
similar to that of the combined width of the two β-strands.
difference in the use of “domain” in structural and molec-
It also provides a hydrophobic core. The dimensions of the
ular biology, since in the latter the term usually indicates a
βαβ motif explain why large parallel sheets that are built
linear section of DNA that appears to influence a biolog-
with this motif always have α-helices on both sides since
ical property where as in structural biology it represents
there is insufficient space on one side of a sheet to accom-
an three-dimensional entity.
modate all of the connecting helices.
A variety of αα motifs are found in proteins depending
on whether the α-helices are in contact with each other D. Protein Folds
after the connecting loop. In cases where the α-helices are
Structural studies on proteins have uncovered a very wide
in contact they are typically inclined at an angle of either
variety of protein folds. At this time the upper limit of the
◦
20 or 50 reflecting the optimal ways to interdigitate side
chains at their intersection. Both types of interaction are number of unique ways in which proteins can fold is un-
abundant in proteins and give rise to parallel or crossed known; however, genomic sequencing has provided a limit
helical bundles. There are also many important examples for the maximum number of folds that might be needed for
of αα motifs where the connections between the two he- the life of an organism by providing an upper limit to the
lices are longer to create a ligand binding site. Important number of proteins in the genome. Fortunately, the number
examples of this type of motif are the helix–turn–helix mo- of unique folds is likely to be considerably less than the
total number of proteins since many proteins of dissimilar
tifs found in calcium binding proteins and DNA binding
function have been found to contain the same fold.
proteins.
The assortment of protein folds observed thus far, at
first glance, appears bewilderingly complex. Careful anal-
C. Domains
ysis of the common structural and topological features
The tertiary structure of a protein describes the manner of these structures has lead to a classification of pro-
in which the secondary structural elements are arranged tein folds according to the content and arrangement of