Page 174 - Academic Press Encyclopedia of Physical Science and Technology 3rd BioChemistry
P. 174
P1: GPAFinal Pages
Encyclopedia of Physical Science and Technology EN013D-616 July 27, 2001 12:5
214 Protein Structure
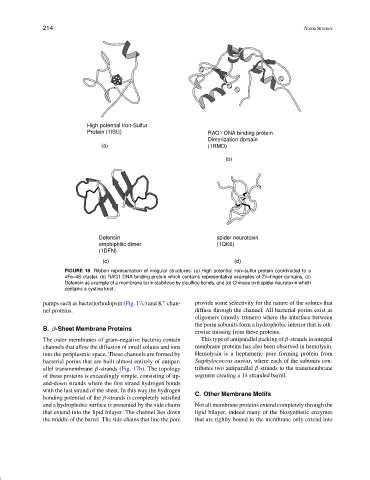
High potential Iron-Sulfur
Protein (1ISU) RAG1 DNA binding protein
Dimerization domain
(a) (1RMD)
(b)
Defensin spider neurotoxin
amphiphilic dimer (1QK6)
(1DFN)
(c) (d)
FIGURE 16 Ribbon representation of irregular structures: (a) High potential iron–sulfur protein coordinated to a
4Fe–4S cluster, (b) RAG1 DNA binding protein which contains representative examples of Zn–finger domains, (c)
Defensin as example of a membrane toxin stabilized by disulfide bonds, and (d) Chinese bird spider neurotoxin which
contains a cystine knot.
+
pumps such as bacteriorhodopsin (Fig. 17c) and K chan- provide some selectivity for the nature of the solutes that
nel proteins. diffuse through the channel. All bacterial porins exist as
oligomers (mostly trimers) where the interface between
the porin subunits form a hydrophobic interior that is oth-
B. β-Sheet Membrane Proteins
erwise missing from these proteins.
The outer membranes of gram-negative bacteria contain This type of antiparallel packing of β-strands in integral
channels that allow the diffusion of small solutes and ions membrane proteins has also been observed in hemolysin.
into the periplasmic space. These channels are formed by Hemolysin is a heptameric pore forming protein from
bacterial porins that are built almost entirely of antipar- Staphylococcus aureus, where each of the subunits con-
allel transmembrane β-strands (Fig. 17b). The topology tributes two antiparallel β-strands to the transmembrane
of these proteins is exceedingly simple, consisting of up- segment creating a 14 stranded barrel.
and-down strands where the first strand hydrogen bonds
with the last strand of the sheet. In this way the hydrogen
C. Other Membrane Motifs
bonding potential of the β-strands is completely satisfied
and a hydrophobic surface is presented by the side chains Not all membrane proteins extend completely through the
that extend into the lipid bilayer. The channel lies down lipid bilayer, indeed many of the biosynthetic enzymes
the middle of the barrel. The side chains that line the pore that are tightly bound to the membrane only extend into