Page 197 - Academic Press Encyclopedia of Physical Science and Technology 3rd BioChemistry
P. 197
P1: GPB Final Pages
Encyclopedia of Physical Science and Technology EN013D-617 July 27, 2001 11:42
Protein Synthesis 237
˚
The structures now available at 5–8 A resolution allow
differentiation of protein α-helices from double-helical
regions of rRNA based on their structural characteristics;
some protein β-sheets can also be seen. The structures
of several individual ribosomal proteins have been solved
by X-ray crystallography or nuclear magnetic resonance
spectroscopy, and the X-ray data on the whole 50S parti-
cle have been correlated with the previously determined
structuresofindividualproteins.Asexpected,theaccumu-
lateddatafromIEM,cross-linking,andneutrondiffraction
helped to position these protein structures within the ribo-
some electron density and will continue to direct place-
ment of proteins and specific regions of rRNA.
Although the locations of many proteins and thousands
of nucleotides must still be teased out of the electron den-
sity maps, structural features of specific regions of the ri-
bosome have already provided mechanistic clues. A more
detailed understanding of the mechanism of protein syn-
thesis will emerge from these structural studies.
E. Mechanistic Clues from Structure
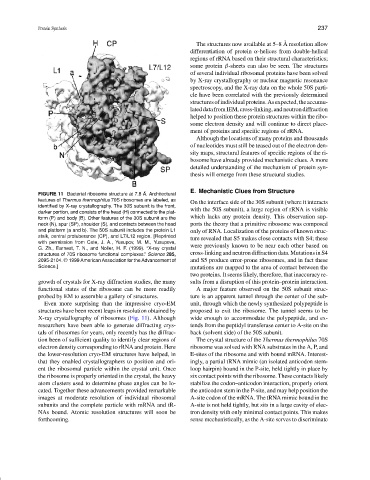
FIGURE 11 Bacterial ribosome structure at 7.8 ˚ A. Architectural
features of Thermus thermophilus 70S ribosomes are labeled, as
On the interface side of the 30S subunit (where it interacts
identified by X-ray crystallography. The 30S subunit is the front,
with the 50S subunit), a large region of rRNA is visible
darker portion, and consists of the head (H) connected to the plat-
which lacks any protein density. This observation sup-
form (P) and body (B). Other features of the 30S subunit are the
neck (N), spur (SP), shoulder (S), and contacts between the head ports the theory that a primitive ribosome was composed
and platform (a and b). The 50S subunit includes the protein L1 only of RNA. Localization of the proteins of known struc-
stalk, central protuberance (CP), and L7/L12 region. [Reprinted
ture revealed that S5 makes close contacts with S4; these
with permission from Cate, J. A., Yusupov, M. M., Yusupova,
G. Zh., Earnest, T. N., and Noller, H. F. (1999). “X-ray crystal were previously known to be near each other based on
structures of 70S ribosome functional complexes.” Science 285, cross-linking and neutron diffraction data. Mutations in S4
2095-2104. © 1999 American Association for the Advancement of and S5 produce error-prone ribosomes, and in fact these
Science.] mutations are mapped to the area of contact between the
two proteins. It seems likely, therefore, that inaccuracy re-
growth of crystals for X-ray diffraction studies, the many sults from a disruption of this protein–protein interaction.
functional states of the ribosome can be more readily A major feature observed on the 50S subunit struc-
probed by EM to assemble a gallery of structures. ture is an apparent tunnel through the center of the sub-
Even more surprising than the impressive cryo-EM unit, through which the newly synthesized polypeptide is
structures have been recent leaps in resolution obtained by proposed to exit the ribosome. The tunnel seems to be
X-ray crystallography of ribosomes (Fig. 11). Although wide enough to accommodate the polypeptide, and ex-
researchers have been able to generate diffracting crys- tends from the peptidyl transferase center to A-site on the
tals of ribosomes for years, only recently has the diffrac- back (solvent side) of the 50S subunit.
tion been of sufficient quality to identify clear regions of The crystal structure of the Thermus thermophilus 70S
electron density corresponding to rRNA and protein. Here ribosome was solved with RNA substrates in the A, P, and
the lower-resolution cryo-EM structures have helped, in E-sites of the ribosome and with bound mRNA. Interest-
that they enabled crystallographers to position and ori- ingly, a partial tRNA mimic (an isolated anticodon stem-
ent the ribosomal particle within the crystal unit. Once loop hairpin) bound in the P-site, held tightly in place by
the ribosome is properly oriented in the crystal, the heavy six contact points with the ribosome. These contacts likely
atom clusters used to determine phase angles can be lo- stabilize the codon–anticodon interaction, properly orient
cated. Together these advancements provided remarkable the anticodon stem in the P-site, and may help position the
images at moderate resolution of individual ribosomal A-site codon of the mRNA. The tRNA mimic bound in the
subunits and the complete particle with mRNA and tR- A-site is not held tightly, but sits in a large cavity of elec-
NAs bound. Atomic resolution structures will soon be tron density with only minimal contact points. This makes
forthcoming. sense mechanistically, as the A-site serves to discriminate